Covalent Inhibition of the Peptidyl-Prolyl Isomerase Pin1 by Sulfopin Results in a Broad Impact on the Phosphoproteome of Human Osteosarcoma U2-OS Cells
- PMID: 40546013
- PMCID: PMC12284463
- DOI: 10.1002/pmic.13980
Covalent Inhibition of the Peptidyl-Prolyl Isomerase Pin1 by Sulfopin Results in a Broad Impact on the Phosphoproteome of Human Osteosarcoma U2-OS Cells
Abstract
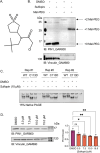
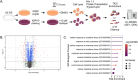
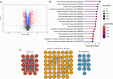
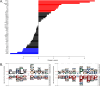
Peptidyl-prolyl isomerase, NIMA-interacting protein 1-(Pin1) catalyses the cis-trans interconversion of the inflexible bond between serine or threonine residues and proline at the +1 position (pSer/pThr-Pro). Although initially discovered as an essential regulator of cell division, Pin1 has since been identified as a regulator of many biological processes and is associated with numerous malignancies and neurodegenerative disorders. Pin1 has been shown to influence phosphorylation by modulating phosphatase accessibility. However, it can also indirectly regulate phosphorylation by isomerizing peptidyl-prolyl bonds on kinases, affecting their subcellular localization and/or substrate specificity. Here, SILAC-based mass spectrometry was employed to identify proteomic and phosphoproteomic changes in human osteosarcoma human osteosarcoma cell line (U2-OS) cells in response to treatment with the selective covalent Pin1 inhibitor Sulfopin. We confirmed that Sulfopin covalently binds Pin1 and profiled Pin1-dependent changes to the proteome and phosphoproteome, identifying 803 phosphosites that underwent significant Sulfopin-dependent changes. The identified phosphosites include substrates for a number of distinct kinases, including protein kinase B (AKT1), aurora kinase A (AURKA), cyclin-dependent kinase (CDK)1 and CK2. Overall, this study reveals the broad impact of Sulfopin on the phosphoproteome, improving our understanding of how Pin1 modulates complex regulatory kinase networks in living cells. SUMMARY: The peptidyl-prolyl isomerase (PPIase) Pin1 has emerged as a potential therapeutic target for numerous malignancies and neurodegenerative disorders based on its altered expression in several diseases. As the activity of Pin1 is phosphorylation-dependent, it is intimately involved with constituents of regulatory kinase networks within cells. To elucidate how Pin1 orchestrates regulatory signalling within cells, we performed quantitative proteomic and phosphoproteomic profiling of SILAC-labelled human osteosarcoma U2-OS cells treated with Sulfopin, a highly selective covalent Pin1 inhibitor. In addition to demonstrating that Pin1 inhibition alters the abundance and phosphorylation of proteins involved in a variety of fundamental cellular processes, these studies revealed that Pin1 inhibition modulates the phosphorylation of 803 phosphorylation sites, ultimately improving our understanding of how a PPIase regulates phosphorylation networks in complex biological systems.
Keywords: CK2; Pin1; Sulfopin; phosphoproteomics; protein kinase; proteomics.
© 2025 The Author(s). PROTEOMICS published by Wiley‐VCH GmbH.
Conflict of interest statement
Xiao Zhen Zhou and Kun Ping Lu are authors of Pin1‐related patents and patent applications. The other authors declare no conflicts of interest.
Figures
References
-
- Lu K. P., Hanes S. D., and Hunter T., “A Human Peptidyl‐Prolyl Isomerase Essential for Regulation of Mitosis,” Nature 380 (1996): 544–547. - PubMed
-
- Yeh E. S. and Means A. R., “PIN1, the Cell Cycle and Cancer,” Nature Reviews Cancer 7 (2007): 381–388. - PubMed
-
- Winkler K. E., Swenson K. I., Kornbluth S., and Means A. R., “Requirement of the Prolyl Isomerase Pin1 for the Replication Checkpoint,” Science 287 (2000): 1644–1647. - PubMed
-
- Van Drogen F., Sangfelt O., Malyukova A., et al., “Ubiquitylation of Cyclin E Requires the Sequential Function of SCF Complexes Containing Distinct hCdc4 Isoforms,” Molecular Cell 23 (2006): 37–48. - PubMed
MeSH terms
Substances
Grants and funding
- 37854/CAPMC/ CIHR/Canada
- 192138/CAPMC/ CIHR/Canada
- 148605/CAPMC/ CIHR/Canada
- 196011/CAPMC/ CIHR/Canada
- 06462/Natural Sciences and Engineering Research Council of Canada
- 1240/Ontario Institute for Cancer Research
- Ontario Graduate Scholarship from the Province of Ontario
- 43257/Canada Foundation for Innovation
- 43822/Canada Foundation for Innovation
- 28946/Canada Foundation for Innovation
- 37854/CAPMC/ CIHR/Canada
- 192138/CAPMC/ CIHR/Canada
- 148605/CAPMC/ CIHR/Canada
- 196011/CAPMC/ CIHR/Canada
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

