MYC/TET3-Regulated TMEM65 Activates OXPHOS-SERPINB3 Pathway to Promote Progression and Cisplatin Resistance in Triple-Negative Breast Cancer
- PMID: 40546127
- PMCID: PMC12442607
- DOI: 10.1002/advs.202500421
MYC/TET3-Regulated TMEM65 Activates OXPHOS-SERPINB3 Pathway to Promote Progression and Cisplatin Resistance in Triple-Negative Breast Cancer
Abstract
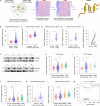
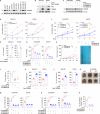
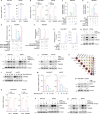
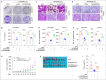
Triple-negative breast cancer (TNBC) is the most lethal subtype of breast cancer due to its aggressive clinical features and the lack of effective targeted therapeutics. Mitochondrial metabolism is intimately linked to TNBC progression and therapeutic resistance and is an attractive therapeutic target for TNBC. Here, it is first reported that human transmembrane protein 65 (TMEM65), a poorly characterized mitochondrial inner-membrane protein-encoding gene in human cancer, acts as a novel oncogene in TNBC to promote tumor growth, metastasis, and cisplatin resistance both in vivo and in vitro. Transcription factor MYC and DNA demethylase ten-eleven translocation 3 (TET3) coordinately upregulate TMEM65 in TNBC, and its upregulation is associated with poor patient survival. Moreover, pharmacological inhibition or knockdown of MYC and TET3 attenuates TMEM65-driven TNBC progression. Mechanistic investigations reveal that TMEM65 enhances mitochondrial oxidative phosphorylation and its byproduct reactive oxygen species (ROS) production. Increased ROS induces the expression of hypoxia-inducible factor 1α (HIF1α), which in turn transcriptionally activates serpin family B member 3 (SERPINB3) to enhance TNBC stemness, thus leading to TNBC progression and cisplatin resistance. Collectively, these findings identify TMEM65 as a vital oncogene of TNBC, unveil its regulatory mechanisms, and shed light on its potential role in TNBC therapy.
Keywords: cancer stemness; chemoresistance; mitochondrial metabolism; transmembrane protein; triple‐negative breast cancer.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Foulkes W. D., Smith I. E., Reis‐Filho J. S., N. Engl. J. Med. 2010, 363, 1938. - PubMed
-
- Poggio F., Bruzzone M., Ceppi M., Pondé N. F., La Valle G., Del Mastro L., de Azambuja E., Lambertini M., Ann. Oncol. 2018, 29, 1497. - PubMed
-
- Zhu Y., Hu Y., Tang C., Guan X., Zhang W., Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188678. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
