A machine learning model for robust prediction of sepsis-induced coagulopathy in critically ill patients with sepsis
- PMID: 40546281
- PMCID: PMC12179180
- DOI: 10.3389/fcimb.2025.1579558
A machine learning model for robust prediction of sepsis-induced coagulopathy in critically ill patients with sepsis
Abstract
Introduction: Sepsis-induced coagulopathy (SIC) is a common disease in patients with sepsis. It denotes higher mortality rates and a poorer prognosis in these patients. This study aimed to develop a practical machine learning (ML) model for the prediction of the risk of SIC in critically ill patients with sepsis.
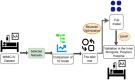
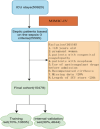
Methods: In this retrospective cohort study, patients were extracted from the Medical Information Mart for Intensive Care IV (MIMIC-IV) database and the Inner Mongolia Autonomous Region People's Hospital database. Sepsis and SIC were defined based on the Sepsis-3 criteria and the criteria developed based on the International Society of Thrombosis and Haemostasis (ISTH), respectively. We compared nine ML models using the Sequential Organ Failure Assessment (SOFA) score in terms of SIC prediction ability. Optimal model selection was based on the superior performance metrics exhibited by the model on the training dataset, the internal validation dataset, and the external validation dataset.
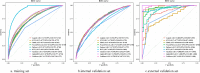
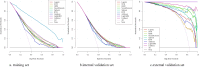
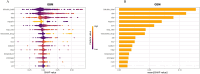
Results: Of the 15,479 patients in MIMIC-IV included in the final cohort, a total of 6,036 (38.9%) patients developed SIC during sepsis. We selected 17 features to construct ML prediction models. The gradient boosting machine (GBM) model was deemed optimal as it achieved high predictive accuracy and reliability across the training, internal, and external validation datasets. The areas under the curve of the GBM model were 0.773 (95%CI = 0.765-0.782) in the training dataset, 0.730 (95%CI = 0.715-0.745) in the internal validation dataset, and 0.966 (95%CI = 0.938-0.994) in the external validation dataset. The Shapley Additive Explanations (SHAP) values illustrated the prediction results, indicating that total bilirubin, red cell distribution width (RDW), systolic blood pressure (SBP), heparin, and blood urea nitrogen (BUN) were risk factors for progression to SIC in patients with sepsis.
Conclusions: We developed an optimal and operable ML model that was able to predict the risk of SIC in septic patients better than the SOFA scoring models.
Keywords: machine learning; predict; risk factor; sepsis; sepsis-induced coagulopathy.
Copyright © 2025 Sun, Zhang, Gong, Ma, Wu, Wu and Siri.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

