Advancing butterfly systematics through genomic analysis
- PMID: 40546317
- PMCID: PMC12180588
- DOI: 10.5281/zenodo.15531392
Advancing butterfly systematics through genomic analysis
Abstract
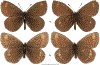
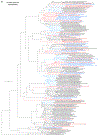
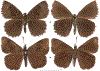
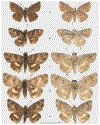
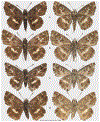
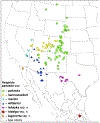
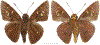
Within the framework of an ongoing comparative genomic study of global butterfly diversity, we construct phylogenetic trees combining all protein-coding genes assembled from the whole genome shotgun data. When viewed in the context of current taxonomy and phenotypic knowledge, the genome-wide phylogeny points to further advances in butterfly systematics, which are presented here. We assign major clades with comparable levels of genetic divergence to the same taxonomic rank and apply criteria involving relative population divergence and gene flow to define species boundaries. As a result, one genus, 13 subgenera, 62 species, and five subspecies are proposed as new (type species in original combinations or type localities are given in parentheses): a genus Ajenorix Grishin, gen. n. (Rapala hypargyria Elwes, 1893) in Deudorigini Doherty, 1886, Lycaenidae [Leach], [1815]; subgenera: in Riodinidae Grote, 1895 (1827): Ouida Grishin, subgen. n. (Dodona ouida Hewitson, 1866) and Egeona Grishin, subgen. n. (Taxila egeon Westwood, 1851) of Dodona Hewitson, 1861, Locris Grishin, subgen. n. (Lasaia oileus Godman, 1903) of Lasaia H. Bates, 1868, Lucispila Grishin, subgen. n. (Hesperia lucianus Fabricius, 1793) of Parvospila J. Hall, 2018, Byzia Grishin, subgen. n. (Lemonias byzeres Hewitson, 1872) of Zelotaea H. Bates, 1868, Arichlosyne Grishin, subgen. n. (Apodemia ochracea Mengel, 1902) of Aricoris Westwood, 1851, and Lenca Grishin, subgen. n. (Lemonias lencates Hewitson, 1875) of Pachythone H. Bates, 1868; in Lycaenidae: Afrix Grishin, subgen. n. (Dipsas antalus Hopffer, 1855) of Capys Hewitson, 1865, Wacus Grishin, subgen. n. (Myrina epirus C. Felder, 1860) of Deudorix Hewitson, 1863, and Crates Grishin, subgen. n. (Hesperia isocrates Fabricius, 1793) of Virachola F. Moore, 1881; and in Hesperiidae Latreille, 1809: Ochloba Grishin, subgen. n. (Poanes batesi Bell, 1935), Ochlata Grishin, subgen. n. (Hesperia venata Bremer & Grey, 1853), and Ochluma Grishin, subgen. n. (Hesperia yuma W. H. Edwards, 1873) of Ochlodes Scudder, 1872; species: in Riodinidae: Lasaia cola Grishin, sp. n. (Mexico: Colima), Curvie wing Grishin, sp. n. (USA: Texas), Curvie westwing Grishin, sp. n. (Mexico: Sonora), Curvie chiapensis Grishin, sp. n. (Mexico: Chiapas), Emesis (Tenedia) tinia Grishin, sp. n. (Argentina), Emesis (Tenedia) guaya Grishin, sp. n. (Uruguay), Emesis (Aphacitis) bugaba Grishin, sp. n. (Panama: Chiriquí), and Synargis rectanga Grishin, sp. n. (Peru: San Martin) and in Hesperiidae: Phanus ecutinus Grishin, sp. n. (Ecuador: Pichincha), Entheus zeus Grishin, sp. n. (Brazil: Amazonas), Entheus guyaneus Grishin, sp. n. (Guyana), Entheus colombeus Grishin, sp. n. (eastern Colombia), Entheus proxemus Grishin, sp. n. (Brazil: Pará), Entheus peruveus Grishin, sp. n. (Peru: Madre de Dios), Entheus hyponota Grishin, sp. n. (Brazil: Amazonas), Entheus lina Grishin, sp. n. (Brazil: Pará), Entheus guato Grishin, sp. n. (Mexico: Chiapas), Entheus pano Grishin, sp. n. (Panama: Darién), Entheus venezuelius Grishin, sp. n. (Venezuela: Aragua), Entheus ecuadius Grishin, sp. n. (Ecuador: Napo), Entheus bogoteus Grishin, sp. n. (Colombia: Bogotá), Cecropterus (Thorybes) notochlorothrix Grishin, sp. n. (Brazil: Santa Catarina), Urbanus (Urbanoides) elma Grishin, sp. n. (Venezuela: Merida), Telegonus (Rhabdoides) missionus Grishin, sp. n. (USA: Texas), Telegonus (Rhabdoides) panavenus Grishin, sp. n. (Panama: Panamá), Telegonus (Rhabdoides) pacificus Grishin, sp. n. (Peru: Piura), Telegonus (Rhabdoides) amazonicus Grishin, sp. n. (Brazil: Rondônia), Telegonus (Rhabdoides) pallidus Grishin, sp. n. (Panama: Darién), Telegonus (Rhabdoides) subfuscus Grishin, sp. n. (Brazil: Santa Catarina), Telegonus (Rhabdoides) elorianus Grishin, sp. n. (likely Southeast or South Brazil), Telegonus (Rhabdoides) perumazon Grishin, sp. n. (Peru: Madre de Dios), Telegonus (Rhabdoides) steinhauseri Grishin, sp. n. (Mexico: Veracruz), Telegonus (Rhabdoides) chiapus Grishin, sp. n. (Mexico: Chiapas), Telegonus (Rhabdoides) colotrix Grishin, sp. n. (Colombia: Cauca), Telegonus (Rhabdoides) flavimargo Grishin, sp. n. (Costa Rica: Limón), Telegonus (Rhabdoides) sobrasus Grishin, sp. n. (Brazil: Santa Catarina), Telegonus (Rhabdoides) chuchuvianus Grishin, sp. n. (Ecuador: Esmeraldas), Telegonus (Rhabdoides) panamus Grishin, sp. n. (Panama: Panamá Oeste), Telegonus (Rhabdoides) tatus Grishin, sp. n. (Panama: Panamá), Telegonus (Rhabdoides) fulvimargo Grishin, sp. n. (Peru: Cuzco), Telegonus (Rhabdoides) alardinus Grishin, sp. n. (Brazil: Rio de Janeiro), Pellicia (Hemipteris) cina Grishin, sp. n. (Brazil: Rondônia), Gorgopas trochicuz Grishin, sp. n. (Peru: Cuzco), Gorgopas trocha Grishin, sp. n. (Colombia: Tolima), Gorgopas trochitango Grishin, sp. n. (Argentina: Salta), Perus (Perus) perus Grishin, sp. n. (Peru: Amazonas), Gomalia westafra Grishin, sp. n. (Ghana: Oti), Chirgus (Chirgus) argentinus Grishin, sp. n. (Argentina: Jujuy), Chirgus (Chirgus) teres Grishin, sp. n. (Peru: Junín), Chirgus (Chirgus) sombrus Grishin, sp. n. (Peru: Puno), Zopyrion (Zopyrion) xerxes Grishin, sp. n. (Honduras: San Pedro Sula), Anisochoria bacchoides Grishin, sp. n. (El Salvador: La Libertad), Onespa nuba Grishin, sp. n. (Mexico: Oaxaca), Vacerra tama Grishin, sp. n. (Mexico: Tamaulipas), Vacerra saltina Grishin, sp. n. (Argentina: Salta), Vacerra cuza Grishin, sp. n. (Peru: Cuzco), Oligoria (Oligoria) tinalandia Grishin, sp. n. (Ecuador: Santo Domingo de los Tsáchilas), Eutychide trombella Grishin, sp. n. (Costa Rica: Heredia), Talides hispina Grishin, sp. n. (Ecuador: Napo), Damas honduras Grishin, sp. n. (Honduras: San Pedro Sula), Damas kenos Grishin, sp. n. (Peru: Loreto), and Damas lavandas Grishin, sp. n. (Peru: Madre de Dios); and subspecies: in Nymphalidae Rafinesque, 1815: Erebia (Erebia) pawloskii bilibinia Grishin, ssp. n. (Russia: Chukotka) and in Hesperiidae: Telegonus (Rhabdoides) alector ecuadoricus Grishin, ssp. n. (Ecuador: Esmeraldas), Hesperia pahaska tehaska Grishin, ssp. n. (USA: Texas), Hesperia pahaska hidalgo Grishin, ssp. n. (Mexico: Hidalgo), and Hesperia pahaska bajanorta Grishin, ssp. n. (Mexico: Baja California Norte). The following are valid genera: Uraneis Bates, 1868, stat. rest. (not Thisbe Hübner, [1819]) and Virachola F. Moore, 1881, stat. rest. (not Deudorix Hewitson, 1863). The following are valid subgenera, not genera or synonyms: Balonca F. Moore, 1901, stat. rest. of Dodona Hewitson, 1861, Ariconias J. Hall & Harvey, 2002, stat. nov. of Aricoris Westwood, 1851, Thisbe Hübner, [1819], stat. nov. of Lemonias Hübner, [1807], and Pseudonymphidia Callaghan, 1985, stat. nov. of Pachythone H. Bates, 1868. The following are valid species, not subspecies or synonyms: Erebia (Erebia) pawloskii Ménétriés, 1859, stat. conf. (not Erebia (Erebia) theano (Tauscher, 1806) or Erebia (Erebia) stubbendorfii Ménétriés, 1847), Erebia (Erebia) demmia B. Warren, 1936, stat. nov. (not Erebia (Erebia) pawloskii Ménétriés, 1859), Lasaia oaxacensis Grishin, 2024, stat. nov. (not Lasaia sessilis Schaus, 1890), Curvie yucatanensis (Godman & Salvin, 1886), stat. rest. (not Curvie emesia (Hewitson, 1867)), Entheus talaus (Linnaeus, 1763), stat. rest. and Entheus pralina Evans, 1952, stat. nov. (not Entheus priassus (Linnaeus, 1758)), Entheus dius Mabille, 1898, stat. rest., Entheus aequatorius Mabille & Boullet, 1919, stat. rest., Entheus latifascius M. Hering, 1925, stat. rest., and Entheus marmato Salazar & Vargas, [2017], stat. nov. (not Entheus matho Godman & Salvin, 1879), Cecropterus (Thorybes) coxeyi (Williams, 1931), stat. rest. (not Cecropterus (Thorybes) egregius (Butler, 1870)), Cecropterus (Thorybes) chlorothrix (Röber, 1925), stat. rest. (not Cecropterus (Thorybes) virescens (Mabille, 1877)), Telegonus (Rhabdoides) hopfferi (Plötz, 1881), stat. rest. (not Telegonus (Rhabdoides) alector (C. Felder & R. Felder, 1867)), Telegonus (Rhabdoides) gilberti (H. Freeman, 1969), stat. rest. (not Telegonus (Rhabdoides) hopfferi (Plötz, 1881), stat. rest), Telegonus (Rhabdoides) bifascia (Herrich-Schäffer, 1869), stat. conf. and Telegonus (Rhabdoides) tinda (Evans, 1952), stat. conf. (not Telegonus (Rhabdoides) latimargo (Herrich-Schäffer, 1869)), Telegonus (Rhabdoides) parmenides (Stoll, 1781), stat. rest., Telegonus (Rhabdoides) crana (Evans, 1952), stat. nov., and Telegonus (Rhabdoides) cyprus (Evans, 1952), stat. nov. (not Telegonus (Rhabdoides) creteus (Cramer, 1780)), Telegonus (Rhabdoides) erana (Evans, 1952), stat. nov. and Telegonus (Rhabdoides) meretrix (Hewitson, 1876), stat. rest. (not Telegonus (Rhabdoides) chiriquensis Staudinger, 1875), Telegonus (Rhabdoides) grullus (Mabille, 1888), stat. rest. (not Telegonus (Rhabdoides) latimargo (Herrich-Schäffer, 1869)), Pellicia (Hemipteris) meno (Mabille, 1889), stat. rest. and Pellicia (Hemipteris) zamia (Plötz, 1882), stat. rest. (not Pellicia (Pellicia) dimidiata Herrich-Schäffer, 1870), Pellicia (Hemipteris) fumida Mabille, 1889, stat. rest., Pellicia (Hemipteris) aequatoria Williams & Bell, 1939, stat. rest., and Pellicia (Hemipteris) toza Evans, 1953, stat. nov. (not Pellicia (Hemipteris) tyana Plötz, 1882), Pellicia (Hemipteris) naja Steinhauser, 1989, stat. nov. (not Pellicia vecina Schaus, 1902, syn. nov.), Gomalia litoralis Swinhoe, 1885, stat. rest. (not Gomalia albofasciata F. Moore, 1879), Chirgus (Chirgus) biseriatus (Weymer, 1890), stat. rest. (not Chirgus (Chirgus) limbata (Erschoff, 1876)), Chirgus (Chirgus) trisignatus (Mabille, 1875), stat. rest. (not Chirgus (Chirgus) bocchoris (Hewitson, 1874)), Zopyrion (Zopyrion) thyas Evans, 1953, stat. nov. (not Zopyrion (Zopyrion) subvariegata Hayward, 1942), Vacerra cecropterus (Draudt, 1923), stat. rest. (not Vacerra hermesia (Hewitson, 1870)), and Damas corope (Herrich-Schäffer, 1869), stat. rest., Damas cervus (Möschler, 1877), stat. rest., and Damas angulis (Plötz, 1886), stat. rest. (not Damas clavus (Herrich-Schäffer, 1869). The following are valid subspecies, not species or synonyms: Erebia (Erebia) pawloskii sajana Staudinger, 1894 stat. rest. (not Erebia (Erebia) pawloskii pawloskii Ménétriés, 1859), Pellicia (Pellicia) dimidiata brasiliensis R. Williams & E. Bell, 1939, stat. rest. (not a synonym of Pellicia (Hemipteris) meno (Mabille, 1889), stat. rest.), Chirgus (Chirgus) biseriatus barrosi Ureta, 1956, stat. nov., Chirgus (Chirgus) bocchoris cuzcona Draudt, 1923, stat. conf., and Damas angulis ampyx (Mabille, 1891), stat. nov. (not a synonym of Damas clavus (Herrich-Schäffer, 1869)). Phareas serenus Plötz, 1883, syn. rev. is a junior objective synonym of Papilio talaus Linnaeus, 1763. The following are junior subjective synonyms, new or transferred between taxa: Esthemopheles Röber, 1903, syn. rev. of Uraneis Bates, 1868, stat. rest., (not of Thisbe Hübner, [1819]), Eudamus oenander Hewitson, 1876, syn. nov. of Aroma aroma (Hewitson, 1867), Aethilla weymeri Plötz, 1882, syn. rev. of Telegonus (Rhabdoides) chiriquensis Staudinger, 1875, Telegonus fabrici Ehrmann, 1918, syn. rev. of Telegonus (Rhabdoides) latimargo (Herrich-Schäffer, 1869) (not Telegonus (Rhabdoides) alardus (Stoll, 1790), Arteurotia demetrius Plötz, 1882, syn. nov. and Pellicia vecina Schaus, 1902, syn. nov. of Pellicia (Hemipteris) tyana Plötz, 1882, Carystus orope Capronnier, 1874, syn. rev. of Tigasis corope (Herrich-Schäffer, 1869) (not of Damas corope (Herrich-Schäffer, 1869)), Damas woldi Shuey, 2024, syn. nov. of Damas corope (Herrich-Schäffer, 1869), stat. rest., and Thracides polles Godman, 1901, syn. rev. and Perichares tripuncta Draudt, 1923, syn. rev. of Damas angulis ampyx (Mabille, 1891), stat. nov. (not of Damas corope (Herrich-Schäffer, 1869)). Pilodeudorix batikelides (W. Holland, 1920) (not Deudorix Hewitson, 1863) is a new genus-species combination and the following are new species-subspecies combinations: Telegonus (Rhabdoides) bifascia siges Mabille, 1903 (not Telegonus (Rhabdoides) creteus (Cramer, 1780)), Telegonus (Rhabdoides) latimargo aquila Evans, 1952 (not Telegonus (Rhabdoides) alardus (Stoll, 1790), and Gomalia jeanneli levana Benyamini, 1990, (not Gomalia elma (Trimen, 1862)). Lectotypes are designated for 24 taxa: Phareas serenus Plötz, 1883 (the Amazonian region), Peleus aeacus Swainson, 1831 (South America), Entheus matho Godman & Salvin, 1879 (Nicaragua), Eudamus hopfferi Plötz, 1881 (Mexico, likely south-central or southern), Telegonus bifascia (Herrich-Schäffer, 1869) (Brazil), Telegonus chiriquensis Staudinger, 1875 (Panama: Chiriquí), Aethilla weymeri Plötz, 1882 (likely Panama: Chiriquí), Pellicia zamia Plötz, 1882 (likely Venezuela), Pellicia tyana Plötz, 1882 (Brazil: likely São Paulo), Arteurotia demetrius Plötz, 1882 (Brazil: likely Rio de Janeiro), Pellicia violacea Mabille, 1891 (Brazil: likely Rio de Janeiro), Pellicia vecina Schaus, 1902 (Brazil: Rio de Janeiro), Pellicia bilinea Mabille, 1889 (Panama: Chiriquí), Pholisora clytius Godman & Salvin, 1897 (Mexico: Nayarit), Bolla semitincta Dyar, 1924 (Mexico: Colima), Carterocephalus biseriatus Weymer, 1890 (Bolivia), Zopyrion sandace Godman & Salvin, 1896 (Mexico: Guerrero), Goniloba clavus Herrich-Schäffer, 1869 (Southeast and South Brazil), Goniloba corope Herrich-Schäffer, 1869 (the Amazonian region, likely Suriname), Hesperia crataea Hewitson, 1876 (Brazil: Bahia), Proteides cervus Möschler, 1877 (Suriname), Proteides ampyx Mabille, 1891 (Panama: Chiriquí), Thracides polles Godman, 1901 (Panama: Chiriquí), and Carystus orope Capronnier, 1874 (Southeast or South Brazil). Neotypes are designated for six taxa: Papilio priassus Linnaeus, 1758 (Suriname), Papilio talaus Linnaeus, 1763 (the Amazonian region), Papilio peleus Linnaeus, 1763 (French Guiana), Peleus aeacus Swainson, 1831 (French Guiana), Eudamus blasius Plötz, 1881 (Southeast or South Brazil), and Hesperia angulis Plötz, 1886 (Panama: Panama). The type locality of Perichares tripuncta Draudt, 1923 is not in South Brazil, but likely in Panama: Chiriquí, as deduced by genomic comparison. Chlosyne flavula blackmorei Pelham, 2008 and Chlosyne palla sterope (W. H. Edwards, 1870) may be sympatric in British Columbia, Canada, and Lon co Grishin, 2023 is sympatric with Lon ma Grishin, 2023 in Monteverde, Costa Rica. Additional specimens of Cecropterus (Thorybes) viridissimus Grishin, 2023 confirm it as a species-level taxon, and Aethilla toxeus Plötz, 1882 is confirmed as a junior subjective synonym of Cecropterus (Murgaria) albociliatus albociliatus (Mabille, 1877) by further genomic sequencing. The holotype of Cecropterus (Thorybes) oaxacensis Grishin, 2023 is illustrated after being spread. Curiously, Onespa gala (Godman, 1900) and Onespa brockorum Austin & A. Warren, 2009 lack overall genetic differentiation typical of species-level taxa. Furthermore, preliminary taxonomic lists of Entheus Hübner, [1819] and Telegonus (Rhabdoides) Scudder, 1889 (from the clade analyzed in this work) are given. Finally, unless stated otherwise, all subgenera, species, subspecies, and synonyms of mentioned genera, subgenera, and species are transferred together with their parent taxa, and taxa not mentioned in this work remain as previously classified.
Keywords: biodiversity; classification; genomics; phylogeny; taxonomy.
Figures



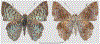
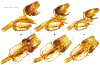


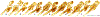



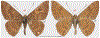
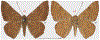




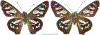

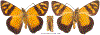



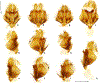


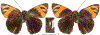

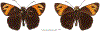


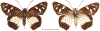


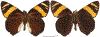


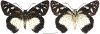
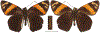


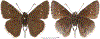

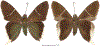







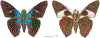

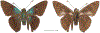
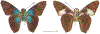
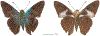


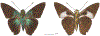
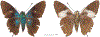
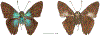

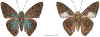


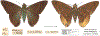
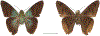
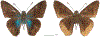
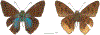
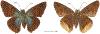
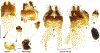
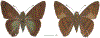
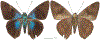
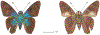

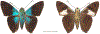

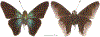








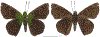
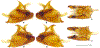
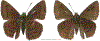

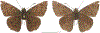







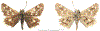
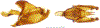
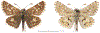
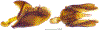
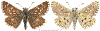
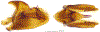
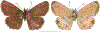
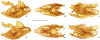
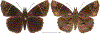
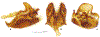

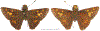
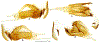

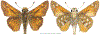
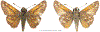
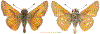





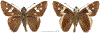

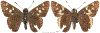

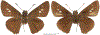
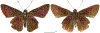





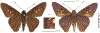
References
-
- Anonymous. 2025. William Swainson (1789-1855) (https://www.museum.zoo.cam.ac.uk/collections-research/collections-uncove...). (Last Accessed 9 May 2025).
-
- Aurivillius POC 1882. Recensio critica Lepidopterorum Musei Ludovicæ Ulricæ qu descripsit Carolus a Linné. Kongliga svenska Vetenskaps-Akademiens Handlingar (Ny Följd) 19(5): 1–188, 1 pl.
-
- Austin GT 1993. A review of the Phanus vitreus group (Lepidoptera: Hesperiidae: Pyrginae). Tropical Lepidoptera 4(suppl. 2): 21–36.
-
- Austin GT 1997. Two new Entheus from Ecuador and Peru (Lepidoptera: Hesperiidae: Pyrginae). Tropical Lepidoptera 8(1): 18–21.
-
- Austin GT, and Mielke OHH. 1998. Hesperiidae of Rondônia, Brazil: Aguna Williams (Pyrginae), with a partial revision and descriptions of new species from Panama, Ecuador, and Brazil. Revista brasileira de Zoologia 14(4): 889–965.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
