Post-transcriptional regulation of cyclin A and B mRNAs by Bruno 1, Cup, and P-bodies
- PMID: 40546942
- PMCID: PMC12179632
- DOI: 10.1016/j.isci.2025.112727
Post-transcriptional regulation of cyclin A and B mRNAs by Bruno 1, Cup, and P-bodies
Abstract
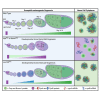
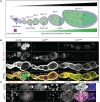
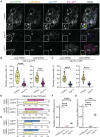
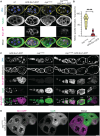
Cell cycle progression relies on tightly regulated Cyclin synthesis and degradation, with Cyclins A and B activating CDK1 to drive mitosis. Dysregulation of Cyclin levels is linked to tumorigenesis, underscoring the importance of studying cyclin mRNA control for cancer therapy development. Using super-resolution microscopy, we show that cyclin A and cyclin B mRNAs associate with Bruno 1 and Cup in nurse cells, and that depletion of either protein disrupts Cyclin expression and reduces mRNA levels. Both mRNAs also accumulate in Me31B-marked P-bodies; however, Me31B selectively affects cyclin B, causing its stage-specific de-repression and decreased stability, while cyclin A remains unaffected. Loss of Me31B enhances cyclin B mRNA's association with Cup, suggesting P-body-independent repression mechanisms. These results highlight the nuanced, mRNA-specific roles of P-body condensates in post-transcriptional regulation, challenging the idea of a uniform, binary mechanism of mRNA repression in P-bodies.
Keywords: Cell biology; Molecular interaction.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


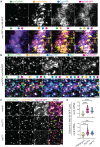
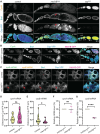
Update of
-
Post-transcriptional regulation of cyclin A and cyclin B mRNAs is mediated by Bruno 1 and Cup, and further fine-tuned within P-bodies.bioRxiv [Preprint]. 2024 Nov 21:2024.10.17.618951. doi: 10.1101/2024.10.17.618951. bioRxiv. 2024. Update in: iScience. 2025 May 21;28(6):112727. doi: 10.1016/j.isci.2025.112727. PMID: 39464095 Free PMC article. Updated. Preprint.
Similar articles
-
Post-transcriptional regulation of cyclin A and cyclin B mRNAs is mediated by Bruno 1 and Cup, and further fine-tuned within P-bodies.bioRxiv [Preprint]. 2024 Nov 21:2024.10.17.618951. doi: 10.1101/2024.10.17.618951. bioRxiv. 2024. Update in: iScience. 2025 May 21;28(6):112727. doi: 10.1016/j.isci.2025.112727. PMID: 39464095 Free PMC article. Updated. Preprint.
-
Molecular mechanisms underlying interferon-alpha-induced G0/G1 arrest: CKI-mediated regulation of G1 Cdk-complexes and activation of pocket proteins.Oncogene. 1999 May 6;18(18):2798-810. doi: 10.1038/sj.onc.1202609. Oncogene. 1999. PMID: 10362250
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Physical exercise training interventions for children and young adults during and after treatment for childhood cancer.Cochrane Database Syst Rev. 2016 Mar 31;3(3):CD008796. doi: 10.1002/14651858.CD008796.pub3. Cochrane Database Syst Rev. 2016. PMID: 27030386 Free PMC article.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
Cited by
-
Trailer Hitch coordinates P-body organization and facilitates transcript-specific mRNA regulation through nuclear actin-mediated feedback loop.bioRxiv [Preprint]. 2025 Jun 20:2025.06.18.660414. doi: 10.1101/2025.06.18.660414. bioRxiv. 2025. PMID: 40611898 Free PMC article. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

