Unraveling the role of GPCR signaling in metabolic reprogramming and immune microenvironment of lung adenocarcinoma: a multi-omics study with experimental validation
- PMID: 40547013
- PMCID: PMC12179119
- DOI: 10.3389/fimmu.2025.1606125
Unraveling the role of GPCR signaling in metabolic reprogramming and immune microenvironment of lung adenocarcinoma: a multi-omics study with experimental validation
Abstract
Background: Lung adenocarcinoma (LUAD) is characterized by metabolic and immune heterogeneity, driving tumor progression and therapy resistance. While G protein-coupled receptors (GPCR) signaling is known to regulate metabolism and immunity in cancers, its role in LUAD remains poorly defined. This study explores the influence of GPCR signaling on LUAD metabolism and immune landscape.
Methods: We performed non-negative matrix factorization (NMF) clustering of GPCR signaling genes in TCGA-LUAD cohort to identify distinct molecular subgroups. A prognostic model was developed based on GPCR signaling genes using least absolute shrinkage and selection operator (LASSO) analysis and Cox regression. Differentially expressed genes were analyzed for metabolic pathway enrichment and immune infiltration. In addition, key genes within GPCR signaling were identified and validated through functional assays.
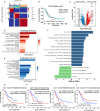
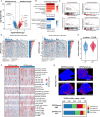
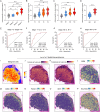
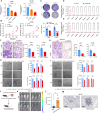
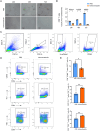
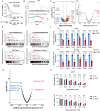
Results: NMF clustering based on GPCR signaling identified three subgroups in LUAD, with cluster 3 exhibiting poorer overall survival and significant enrichment in multiple prognostic associated metabolism pathways including purine, pyrimidine, glyoxylate and dicarboxylate metabolism. Then, we developed a GPCRscore prognostic model and validated across multiple cohorts, which effectively stratified LUAD patients into distinct risk groups. High-risk LUAD patients had an immunosuppressive microenvironment and activated metabolic reprogramming. ADM was identified as a key gene in the high-risk group, correlating with tumor stage, immune suppression, and resistance to immunotherapy. Clinically, ADM was highly expressed in tumor tissues and shows elevated concentrations in the peripheral blood of patients with advanced-stage LUAD. Subsequently, we demonstrated that knock-down of ADM in LUAD cells impaired their proliferation, migration, and invasion, while also reducing the angiogenic potential of endothelial cells in vitro. Adrenomedullin promoted LUAD progression in a murine metastasis model. Further, adrenomedullin inhibited CD8+ T cells proliferation, induced exhaustion, and impaired cytotoxic function. Finally, drug sensitivity and cell viability analysis showed LUAD patients with high levels of ADM exhibited sensitivity to the treatment of Staurosporine and Dasatinib.
Conclusions: In summary, this study reveals the pivotal role of GPCR signaling particularly mediated by ADM in orchestrating metabolic reprogramming and immune modulation in LUAD. ADM emerges as a potential predictive biomarker and therapeutic target, offering valuable implications for optimizing strategies.
Keywords: ADM; GPCR signaling; immune microenvironment; lung adenocarcinoma; metabolic reprogramming; prognostic model.
Copyright © 2025 Wang, Wang, Zhao and Gu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

