Rapid and direct discovery of functional tumor specific neoantigens by high resolution mass spectrometry and novel algorithm prediction
- PMID: 40547114
- PMCID: PMC12179604
- DOI: 10.1016/j.cellin.2025.100251
Rapid and direct discovery of functional tumor specific neoantigens by high resolution mass spectrometry and novel algorithm prediction
Abstract
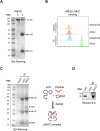
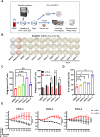
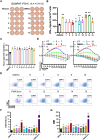
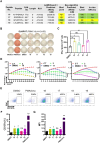
While immune cell therapies have transformed cancer treatment, achieving comparable success in solid tumors remains a significant challenge compared to hematologic malignancies like non-Hodgkin lymphoma (NHL) and multiple myeloma (MM). Over the past four decades, various immunotherapeutic strategies, including tumor vaccines, tumor-infiltrating lymphocyte (TIL) therapies, and T cell receptor (TCR) therapies, have demonstrated clinical efficacy in select solid tumors, suggesting potential advantages over CAR-T and CAR-NK cell therapies in specific contexts. The dynamic nature of the cancer-immunity cycle, characterized by the continuous evolution of tumor-specific neoantigens, enables tumors to evade immune surveillance. This highlights the urgent need for rapid and accurate identification of functional tumor neoantigens to inform the design of personalized tumor vaccines. These vaccines can be based on mRNA, dendritic cells (DCs), or synthetic peptides. In this study, we established a novel platform integrating immunoprecipitation-mass spectrometry (IP-MS) for efficient and direct identification of tumor-specific neoantigen peptides. By combining this approach with our proprietary AI-based prediction algorithm and high-throughput in vitro functional validation, we can generate patient-specific neoantigen candidates within six weeks, accelerating personalized tumor vaccine development.
Keywords: Mass spectrometry; Neoantigen; New algorithm; Tumor vaccine.
© 2025 The Authors.
Conflict of interest statement
The authors declare that there are no conflicts of interest related to this manuscript.
Figures



Similar articles
-
Chimeric antigen receptor (CAR) T-cell therapy for people with relapsed or refractory diffuse large B-cell lymphoma.Cochrane Database Syst Rev. 2021 Sep 13;9(9):CD013365. doi: 10.1002/14651858.CD013365.pub2. Cochrane Database Syst Rev. 2021. PMID: 34515338 Free PMC article.
-
AI-Driven Antimicrobial Peptide Discovery: Mining and Generation.Acc Chem Res. 2025 Jun 17;58(12):1831-1846. doi: 10.1021/acs.accounts.0c00594. Epub 2025 Jun 3. Acc Chem Res. 2025. PMID: 40459283 Free PMC article. Review.
-
Antiemetics for adults for prevention of nausea and vomiting caused by moderately or highly emetogenic chemotherapy: a network meta-analysis.Cochrane Database Syst Rev. 2021 Nov 16;11(11):CD012775. doi: 10.1002/14651858.CD012775.pub2. Cochrane Database Syst Rev. 2021. PMID: 34784425 Free PMC article.
-
Mitigation and Management of Common Toxicities Associated with the Administration of CAR-T Therapies in Oncology Patients.Drug Saf. 2025 Jul;48(7):719-737. doi: 10.1007/s40264-025-01538-5. Epub 2025 Mar 19. Drug Saf. 2025. PMID: 40108072 Free PMC article. Review.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
References
-
- Borno H., Moses K.A., Monk P., Fitch K., Harmon M., Flanders S., Mckay R.R. Sipuleucel-T (sip-T) and oral androgen axis inhibitors in black men being treated for metastatic castration-resistance prostate cancer (mCRPC): Observations from the Medicare Fee for Service (FFS) population. Journal of Clinical Oncology. 2020;38
-
- Chervin A.S., et al. 2021. Bispecific binding molecules. WO2021163366A1.
-
- Chervin A.S., Stone J.D., Konieczna I., Calabrese K.M., Wang N., Haribhai D., Dong F., White M.K., Rodriguez L.E., Bukofzer G.T., Ellis P.A., Cosgrove C., Hecquet C., Clarin J.D., Palma J.P., Reilly E.B. ABBV-184: A novel survivin-specific TCR/CD3 bispecific T-cell engager is active against both solid tumor and hematologic malignancies. Molecular Cancer Therapeutics. 2023;22:903–912. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

