Identification and validation of extracellular matrix-related genes in the progression of gastric cancer with intestinal metaplasia
- PMID: 40547168
- PMCID: PMC12179931
- DOI: 10.4251/wjgo.v17.i6.105160
Identification and validation of extracellular matrix-related genes in the progression of gastric cancer with intestinal metaplasia
Abstract
Background: Gastric cancer (GC) is a highly lethal malignancy with a high incidence and mortality rate globally. Its development follows the Correa model, with intestinal metaplasia (IM) being a critical precursor to GC. However, the mechanisms underlying IM progression to GC remain unclear. This study explored extracellular matrix (ECM)-related gene changes during IM progression to GC, aiming to identify biomarkers that could improve early diagnosis and treatment strategies for GC, ultimately enhancing patient outcomes.
Aim: To analyze transcriptome sequencing data, molecular biomarkers that can predict GC risk and monitor IM progression can be identified, providing new insights and strategies for preventing IM-GC transformation.
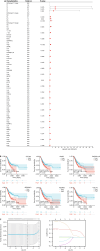
Methods: Weighted gene co-expression network analysis served for confirming gene modules. Upregulated ECM-related genes were further tested using univariate Cox regression and least absolute shrinkage and selection operator analysis to select hub genes and construct a survival analysis model. The intestinal cell model was established by stimulating GES-1 cells with chenodeoxycholic acid.
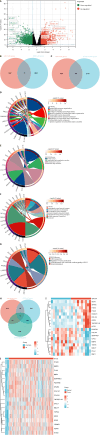
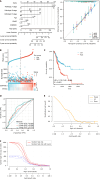
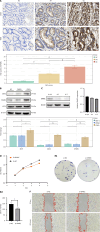
Results: Weighted gene co-expression network analysis identified 1709 differentially expressed genes from the GSE191275 dataset, while The Cancer Genome Atlas stomach adenocarcinoma revealed 4633 differentially expressed genes. The intersection of these datasets identified 71 upregulated and 171 downregulated genes, which were enriched in ECM-related pathways. Univariate Cox regression analysis identified six genes with prognostic significance, and least absolute shrinkage and selection operator regression pinpointed secreted protein acidic and rich in cysteine (SPARC) and SERPINE1 as non-zero coefficient genes. A prognostic model integrating clinical tumor node metastasis staging, age, SERPINE1, and SPARC was constructed. Immunohistochemistry analysis confirmed an increasing expression of SPARC protein from normal gastric mucosa (-), to IM (+- to +), and to GC (+ to ++), with significant differences (P < 0.05). Western blot analysis demonstrated significantly higher SPARC expression in induced intestinal cells compared to GES-1. Furthermore, after SPARC knockdown in the human GC cell line HGC27, cell counting kit-8 and colony formation assays showed a reduction in cell proliferative ability, while the wound healing assay revealed impaired cell migration capacity.
Conclusion: Comprehensive analysis suggested that a model incorporating clinical tumor node metastasis staging, age, and SPARC/SERPINE1 expression served as a prognostic predictor for GC. Moreover, elevated SPARC expression in IM and GC suggests its potential as a proper biomarker to detect GC in early stage and as a novel therapeutic target, guiding clinical applications.
Keywords: SERPINE1; Extracellular matrix; Gastric cancer; Intestinal metaplasia; SPARC.
©The Author(s) 2025. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors declare that they have no conflicts of interest.
Figures




Similar articles
-
Construction and validation of a lipid metabolism-related genes prognostic signature for skin cutaneous melanoma.Biochem Biophys Res Commun. 2025 Aug 15;775:152115. doi: 10.1016/j.bbrc.2025.152115. Epub 2025 May 29. Biochem Biophys Res Commun. 2025. PMID: 40460484
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Molecular feature-based classification of retroperitoneal liposarcoma: a prospective cohort study.Elife. 2025 May 23;14:RP100887. doi: 10.7554/eLife.100887. Elife. 2025. PMID: 40407808 Free PMC article.
-
Comprehensive analysis of anosmin-1 as a potential biomarker and its correlation with epithelial-mesenchymal transition in advanced gastric cancer.3 Biotech. 2025 Jul;15(7):222. doi: 10.1007/s13205-025-04361-y. Epub 2025 Jun 21. 3 Biotech. 2025. PMID: 40546398 Free PMC article.
-
Adenocarcinoma risk in gastric atrophy and intestinal metaplasia: a systematic review.BMC Gastroenterol. 2017 Dec 11;17(1):157. doi: 10.1186/s12876-017-0708-4. BMC Gastroenterol. 2017. PMID: 29228909 Free PMC article.
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. - PubMed
-
- Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396:635–648. - PubMed
-
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209–249. - PubMed
-
- Correa P. Human gastric carcinogenesis: a multistep and multifactorial process--First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. - PubMed
-
- Correa P, Shiao YH. Phenotypic and genotypic events in gastric carcinogenesis. Cancer Res. 1994;54:1941s–1943s. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

