Genomic and phylogeographical analysis revealed CTX-M-55 producing Escherichia coli ST10 and ST2325 clones of One Health concern from dairy farm waste in Gansu, China
- PMID: 40547208
- PMCID: PMC12179727
- DOI: 10.1016/j.onehlt.2025.101101
Genomic and phylogeographical analysis revealed CTX-M-55 producing Escherichia coli ST10 and ST2325 clones of One Health concern from dairy farm waste in Gansu, China
Abstract
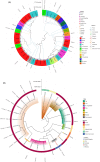
Extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli resists broad-spectrum cephalosporins, considered as a critical priority pathogen, and its presence in animals, humans, and the environment highlights its significance as a One Health issue. Dairy farm waste is a potential environmental contaminant and can serves as a significant reservoir for the emergence and spread of ESBL-producing E. coli, which belongs to One Health clones and poses a serious global threat. This study aimed to investigate the occurrence and genomic characteristics of ESBL-producing E. coli clones of One Health concern from dairy farm waste in Gansu, China. In this study, we isolated and characterized two CTX-M-55 ESBL-producing E. coli strains, ZYX8158 and ZYS8091, which belong to One Health clones. The genomic analysis revealed a large resistome, mobilome, virulome, and plasmidome was acquired by both ESBL-producing E. coli strains. The genome-based typing revealed that E. coli ZYX8158 and ZYS8091 belonged to globally disseminated clones ST10 (O73:H31 serotype) and ST2325 (O66:H25 serotype), respectively. Phylogeographical analysis revealed both strains as potential One Health clones due to their clustering with related E. coli strains isolated from animal, human, and environmental sources, regardless of geographical boundaries, indicating their zoonotic potential and clonal spread in the One Health sector. This study highlights that dairy farm waste can be a potential source of the emergence and dissemination of One Health clones of critical priority ESBL-producing E. coli in One Health settings, which demands continuous and integrated genomic surveillance for comprehensive knowledge and mitigation strategies.
Keywords: ESBL-producing E. coli; Genomic surveillance; Mobilome; One Health clones; Resistome; Virulome.
© 2025 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare no financial and personal competing interests.
Figures

Similar articles
-
Comprehensive regional study of ESBL Escherichia coli: genomic insights into antimicrobial resistance and inter-source dissemination of ESBL genes.Front Microbiol. 2025 Jun 10;16:1595652. doi: 10.3389/fmicb.2025.1595652. eCollection 2025. Front Microbiol. 2025. PMID: 40556893 Free PMC article.
-
Genomic evidences of gulls as reservoirs of critical priority CTX-M-producing Escherichia coli in Corcovado Gulf, Patagonia.Sci Total Environ. 2023 May 20;874:162564. doi: 10.1016/j.scitotenv.2023.162564. Epub 2023 Mar 3. Sci Total Environ. 2023. PMID: 36870482
-
Prevalence of ESBL-producing Escherichia coli in sub-Saharan Africa: A meta-analysis using a One Health approach.One Health. 2025 Jun 2;20:101090. doi: 10.1016/j.onehlt.2025.101090. eCollection 2025 Jun. One Health. 2025. PMID: 40529906 Free PMC article. Review.
-
Emergence of Escherichia coli sequence type 131 clone carrying blaCTX-M genes in Guilan Province, Northern Iran.Microbiol Spectr. 2025 Aug 19:e0227024. doi: 10.1128/spectrum.02270-24. Online ahead of print. Microbiol Spectr. 2025. PMID: 40827951
-
Ecological and public health dimensions of ESBL-producing Escherichia coli in bats: A One Health perspective.Vet World. 2025 May;18(5):1199-1213. doi: 10.14202/vetworld.2025.1199-1213. Epub 2025 May 17. Vet World. 2025. PMID: 40584129 Free PMC article. Review.
References
-
- Shoaib M., He Z., Geng X., Tang M., Hao R., Wang S., Shang R., Wang X., Zhang H., Pu W. The emergence of multi-drug resistant and virulence gene carrying Escherichia coli strains in the dairy environment: a rising threat to the environment, animal, and public health. Front. Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1197579. - DOI - PMC - PubMed
-
- Wang Q., Wang W., Zhu Q., Shoaib M., Chengye W., Zhu Z., Wei X., Bai Y., Zhang J. The prevalent dynamic and genetic characterization of mcr-1 encoding multidrug resistant Escherichia coli strains recovered from poultry in Hebei, China. J. Glob. Antimicrob. Resist. 2024;38:354–362. doi: 10.1016/j.jgar.2024.04.001. - DOI - PubMed
-
- Dong N., Zeng Y., Cai C., Sun C., Lu J., Liu C., Zhou H., Sun Q., Shu L., Wang H., Wang Y., Wang S., Wu C., Chan E.W., Chen G., Shen Z., Chen S., Zhang R. Prevalence, transmission, and molecular epidemiology of tet(X)-positive bacteria among humans, animals, and environmental niches in China: an epidemiological, and genomic-based study. Sci. Total Environ. 2022;818 doi: 10.1016/j.scitotenv.2021.151767. - DOI - PubMed
LinkOut - more resources
Full Text Sources

