Mito-fission gene prognostic model for colorectal cancer
- PMID: 40547309
- PMCID: PMC12182055
- DOI: 10.7717/peerj.19522
Mito-fission gene prognostic model for colorectal cancer
Abstract
Background: Dysregulated cellular metabolism is one of the major causes of colorectal cancer (CRC), including mitochondrial fission. Therefore, this study focuses on the specific regulatory mechanisms of mitochondrial dysfunction on CRC, which will provide theoretical guidance for CRC in the future.
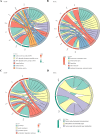
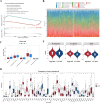
Methods: The Cancer Genome Atlas (TCGA)-CRC dataset, GSE103479 dataset and 40 mitochondrial fission-related genes (MFRGs) were downloaded in this study. The differentially expressed genes (DEGs) were analyzed in TCGA-CRC samples. Using MFRGs scores as traits, key module genes associated with its scores were screened by weighted gene co-expression network analysis (WGCNA). Then, differentially expressed MFRGs (DE-MFRGs) were obtained by intersecting DEGs and key module genes. Next, DE-MFRGs were subjected to univariate Cox, least absolute shrinkage and selection operator (LASSO), multivariate Cox and stepwise regression analysis to scree hub genes and to construct the risk model. The risk model was validated in GSE103479. Finally, the hub genes were comprehensively investigated through a multi-faceted approach encompassing clinical characteristic analysis, Gene Set Enrichment Analysis (GSEA), immune infiltration analysis, and drug sensitivity prediction. Subsequently, the expression levels of the identified key genes were validated utilizing quantitative real-time fluorescence PCR (qRT-PCR), reinforcing the findings and ensuring their accuracy.
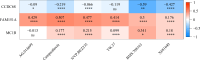
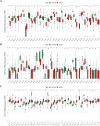
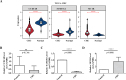
Results: The 49 DE-MFRGs were gained by intersecting 3,310 DEGs and 1,952 key module genes. Then, CCDC68, FAM151A and MC1R were screened as hub genes. Also, the risk model validated in GSE103479 showed that the higher the risk score, the worse the survival of CRC patients. Furthermore, T/N/M stages were differences in risk scores between subgroups of clinical characteristics. The memory CD4+ T cell and plasma cell were more significant differences in the low-risk group samples. The 51 drugs were showed a better response in the high-risk group patients. RT-qPCR validation results showed that CCDC68 and FAM151A were down-regulated in CRC, while MC1R was up-regulated, consistent with the validation set results. And FAM151A and MC1R showed highly significant difference between CRC and normal samples (P < 0.0001).
Conclusion: In this study, we found CCDC68, FAM151A and MC1R as potential hub genes in CRC, and analyzed the molecular mechanism of mitochondrial affecting CRC, which would provide theoretical reference value for CRC.
Keywords: Colorectal cancer; Gene; Mitochondria; Pan-cancer analysis; Risk model.
©2025 Liu et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures








Similar articles
-
Construction of a New Ferroptosis-related Prognosis Model for Survival Prediction in Colorectal Cancer.Curr Med Chem. 2025;32(20):4132-4146. doi: 10.2174/0109298673296767240116215814. Curr Med Chem. 2025. PMID: 38362684
-
Construction and validation of a lipid metabolism-related genes prognostic signature for skin cutaneous melanoma.Biochem Biophys Res Commun. 2025 Aug 15;775:152115. doi: 10.1016/j.bbrc.2025.152115. Epub 2025 May 29. Biochem Biophys Res Commun. 2025. PMID: 40460484
-
Role of lysine acetylation-related genes in the diagnosis and prognosis of glioma.Sci Rep. 2025 Jul 1;15(1):20762. doi: 10.1038/s41598-025-07738-4. Sci Rep. 2025. PMID: 40593136 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
References
-
- Bugajova M, Raudenska M, Hanelova K, Navratil J, Gumulec J, Petrlak F, Vicar T, Hrachovinova S, Masarik M, Kalfert D, Grega M, Plzak J, Betka J, Balvan J. Glutamine and serum starvation alters the ATP production, oxidative stress, and abundance of mitochondrial RNAs in extracellular vesicles produced by cancer cells. Scientific Reports. 2024;14:25815. doi: 10.1038/s41598-024-73943-2. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

