A Fully In Silico Protocol to Understand Olfactory Receptor-Odorant Interactions
- PMID: 40547669
- PMCID: PMC12177607
- DOI: 10.1021/acsomega.4c08181
A Fully In Silico Protocol to Understand Olfactory Receptor-Odorant Interactions
Abstract
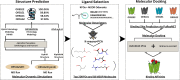
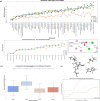
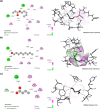
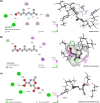
Understanding olfactory receptor (OR)-odorant interaction is crucial for unraveling the molecular intricacies of smell, a sense that is essential for health and survival and has potential therapeutic applications. Nevertheless, the absence of comprehensive experimental data concerning ORs has significantly impeded the understanding of the structural dimensions of olfaction, thereby necessitating innovative approaches to elucidate the structural intricacies of ORs. In this study, we developed an in silico protocol to predict OR structures and study relevant odorant interactions using the OR51E2-propionate complex as a reference. We also developed a hybrid homology modeling strategy leveraging homologous Alphafold structures. This approach resulted in structures with better stability than Alphafold predicted models, as validated through molecular dynamics simulations. Our pipeline accurately replicated experimental findings for OR51E2 and was extended to three homologous ORs: OR51E1, OR51D1, and OR51G2. We used a total of 217 molecules from the M2OR database and key food odorants and applied K-nearest neighbor clustering, selecting a total of 78 representative molecules based on their proximity to cluster centroids for molecular docking studies. Our computational pipeline successfully verified over 25 previously established odorant-OR relationships, including the identification of potential interactions between OR51G2 and molecules such as trans-2-nonenal and acetyl glutamic acid. This framework provides an efficient method for predicting and characterizing potential OR-odorant pairs, streamlining the discovery process prior to experimental confirmation and advancing our understanding of OR binding mechanisms.
© 2025 The Authors. Published by American Chemical Society.
Figures



Similar articles
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
-
Computer and mobile technology interventions for self-management in chronic obstructive pulmonary disease.Cochrane Database Syst Rev. 2017 May 23;5(5):CD011425. doi: 10.1002/14651858.CD011425.pub2. Cochrane Database Syst Rev. 2017. PMID: 28535331 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
References
LinkOut - more resources
Full Text Sources
