Interrogation of macrophage-related prognostic signatures reveals a potential immune-mediated therapy strategy by histone deacetylase inhibition in glioma
- PMID: 40548122
- PMCID: PMC12179195
- DOI: 10.3389/fonc.2025.1554845
Interrogation of macrophage-related prognostic signatures reveals a potential immune-mediated therapy strategy by histone deacetylase inhibition in glioma
Abstract
Background: Glioma-associated macrophages (GAMs) originate from intracranially resident microglia and myeloid-derived macrophages. In the glioma microenvironment, these two types of macrophages tend to adopt a specialized activation state known as type 2 or M2 macrophages and play crucial roles in the progression of glioma.
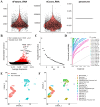
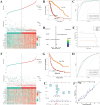
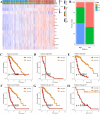
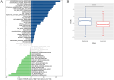
Methods: To identify genes associated with GAMs, we intersected genes identified from single-cell RNA sequencing (scRNA-seq) data (specific to GAMs) with M2 macrophage module genes derived from weighted gene coexpression network analysis (WGCNA). Prognostic genes were screened using univariate Cox regression, multivariate Cox regression, and least absolute shrinkage and selection operator (LASSO) regression analysis. These genes were used to construct and validate prognostic signatures and to delineate the immune landscape. During drug screening, Vorinostat exhibited the highest risk score and the lowest half-maximal inhibitory concentration (IC50). The expression of the 14 prognostic genes was further investigated using a glioma cell-macrophage co-culture model.
Results: Fourteen prognostic genes (TREM2, GAL3ST4, AP1B1, SLA, CYBB, CD53, SLC37A2, ABI3, RIN3, SCIN, SIGLEC10, C3, PLEKHO2, and PLXDC2) were identified. The prognostic model constructed from these genes demonstrated robust predictive efficacy. Based on this model, Vorinostat was prioritized as a candidate therapeutic agent, and subsequent validation confirmed its significant inhibitory effects on the glioma microenvironment.
Conclusion: These findings elucidate the molecular mechanisms of GAMs in glioma, uncover the immunological landscape of the tumor microenvironment, and identify potential therapeutic targets and drug action mechanisms.
Keywords: glioma; glioma-associated macrophages; histone deacetylase inhibitors; immune microenvironment; prognostic signature; vorinostat.
Copyright © 2025 Wang, Wang, Chen, Ding, Fan, Liang, Zhang, Li, Zhang, Yu, Zhang, Li, Li and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Miscellaneous

