LncRNA MEG3/CTCF-CXCR4 axis functions in the regulation of breast cancer cell migration
- PMID: 40548301
- PMCID: PMC12178829
- DOI: 10.1016/j.ncrna.2025.05.014
LncRNA MEG3/CTCF-CXCR4 axis functions in the regulation of breast cancer cell migration
Abstract
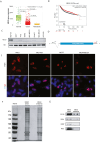
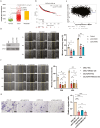
Loss or decreased expression of lncRNA MEG3 is a frequent event in the progression of many different malignancies. Overexpression of MEG3 in breast cancer cell lines MCF7 or MDA-MB-231 prevented cell migration, whereas depletion of MEG3 in human mammary epithelial cell line MCF10A strikingly promoted cell migration. As RNA-protein interactions are vital for RNA to function, RNP assembled on MEG3 in vivo was purified using affinity purification followed by mass spectrometry, which revealed ∼600 proteins with the potential to interact with MEG3. Bioinformatic analysis on RNA-seq data from MCF7 with MEG3 overexpression and MCF10A with MEG3 depletion led to the identification of CXCR4 as the major downstream mediator negatively regulated by MEG3 that facilitated breast cancer cell migration. In addition, the chromatin regulator CTCF emerged as the MEG3-binding protein that might regulate CXCR4 expression after comparison of proteins presenting in MEG3 lncRNP to ChIP-seq data and GPSAdb data of CXCR4. Further evidence was provided to show CTCF upregulated the expression of CXCR4 at transcriptional level, whereas co-expression of MEG3 with CTCF abolished transcriptional activation of CXCR4. Overall, our study pinpoints the importance of MEG3/CTCF-CXCR4 axis in regulating migration of breast cancer cells and provides novel insight into the mechanism of lncRNA MEG3 in cancer development.
Keywords: Breast cancer; CTCF; CXCR4; Cell migration; lncRNA MEG3.
© 2025 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures






Similar articles
-
Long Noncoding RNA MEG3-205/Let-7a/MyD88 Axis Promotes Renal Inflammation and Fibrosis in Diabetic Nephropathy.Kidney Dis (Basel). 2022 Mar 17;8(3):231-245. doi: 10.1159/000523847. eCollection 2022 May. Kidney Dis (Basel). 2022. PMID: 35702702 Free PMC article.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
PARP-1 as a novel target in endocrine-resistant breast cancer.J Exp Clin Cancer Res. 2025 Jun 16;44(1):175. doi: 10.1186/s13046-025-03441-4. J Exp Clin Cancer Res. 2025. PMID: 40518539 Free PMC article.
-
The mechanism of adenosine-mediated activation of lncRNA MEG3 and its antitumor effects in human hepatoma cells.Int J Oncol. 2016 Jan;48(1):421-9. doi: 10.3892/ijo.2015.3248. Epub 2015 Nov 18. Int J Oncol. 2016. PMID: 26647875
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article.
References
-
- Wang Y., Li R.-W., Li X.-T., Xin Y. Decreased long non-coding RNA MEG3 expression is associated with survival outcome and lymph node metastasis: a meta-analysis. Int. J. Clin. Exp. Med. 2017;10(4):5913–5921.
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

