Resident memory T cell development is gradual and shows AP-1 gene expression in mature cells
- PMID: 40548376
- PMCID: PMC12220954
- DOI: 10.1172/jci.insight.187381
Resident memory T cell development is gradual and shows AP-1 gene expression in mature cells
Abstract
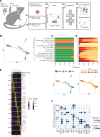
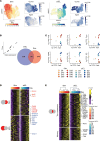
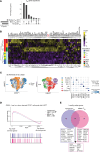
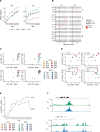
Tissue-resident memory T (TRM) cells play a central role in immune responses across all barrier tissues after infection. However, the mechanisms that drive TRM differentiation and priming for their recall effector function remains unclear. In this study, we leveraged newly generated and publicly available single-cell RNA-seq data generated across 10 developmental time points to define features of CD8+ TRM across both skin and small-intestine intraepithelial lymphocytes (siIEL). We employed linear modeling to capture gene programs that increase their expression levels in T cells transitioning from an effector to a memory state. In addition to capturing tissue-specific gene programs, we defined a temporal TRM signature across skin and siIEL that can distinguish TRM from circulating T cell populations. This TRM signature highlights biology that is missed in published signatures that compared bulk TRM to naive or nontissue resident memory populations. This temporal TRM signature included the AP-1 transcription factor family members Fos, Fosb, Fosl2, and Junb. ATAC-seq analysis detected AP-1-specific motifs at open chromatin sites in mature TRM. Cyclic immunofluorescence (CyCIF) tissue imaging detected nuclear colocalization of AP-1 members in resting CD8+ TRM greater than 100 days after infection. Taken together, these results reveal a critical role of AP-1 transcription factor members in TRM biology.
Keywords: Adaptive immunity; Immunology; Inflammation; T cell development.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

