Integrated Single-Tip IMAC-HILIC Enables Simultaneous Analysis of Plant Phosphoproteomics and N-Glycoproteomics
- PMID: 40548449
- PMCID: PMC12235695
- DOI: 10.1021/acs.jproteome.5c00185
Integrated Single-Tip IMAC-HILIC Enables Simultaneous Analysis of Plant Phosphoproteomics and N-Glycoproteomics
Abstract
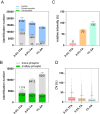
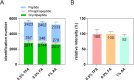
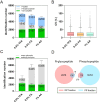
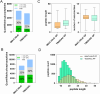
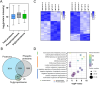
Protein phosphorylation and N-glycosylation are key post-translational modifications (PTMs) in plants that regulate critical signaling processes. However, coanalysis of these PTMs is often complicated by their relatively low abundance and divergent enrichment requirements. Here, we present a single-tip IMAC-HILIC approach that integrates immobilized metal affinity chromatography (IMAC) and hydrophilic interaction chromatography (HILIC) materials within one pipet tip, enabling concurrent enrichment and sequential elution of phosphopeptides and N-glycopeptides. This integrated workflow effectively reduces phosphopeptide coelution during N-glycopeptide elution and streamlines sample processing. In direct comparison with the tandem-tip TIMAHAC method, our single-tip strategy achieves a comparable identification depth and offers superior quantitative accuracy for N-glycopeptides. We further demonstrate its applicability by examining the impact of calcium deprivation in Arabidopsis, revealing distinct global changes in both the phosphoproteome and N-glycoproteome. Our optimized protocol thus provides a straightforward and high-throughput platform for dual PTM profiling in complex plant samples, paving the way for broader investigations of PTM crosstalk in diverse physiological and stress responses.
Keywords: N-glycoproteomics; enrichment; hydrophilic interaction Chromatography; immobilized metal affinity chromatography; phosphoproteomics.
Figures

References
-
- Hsu C. C., Zhu Y., Arrington J. V., Paez J. S., Wang P., Zhu P., Chen I. H., Zhu J. K., Tao W. A.. Universal Plant Phosphoproteomics Workflow and Its Application to Tomato Signaling in Response to Cold Stress. Mol. Cell Proteomics. 2018;17(10):2068–2080. doi: 10.1074/mcp.TIR118.000702. - DOI - PMC - PubMed
-
- Sang T., Chen C. W., Lin Z., Ma Y., Du Y., Lin P. Y., Hadisurya M., Zhu J. K., Lang Z., Tao W. A., Hsu C. C., Wang P.. DIA-Based Phosphoproteomics Identifies Early Phosphorylation Events in Response to EGTA and Mannitol in Arabidopsis. Mol. Cell Proteomics. 2024;23(8):100804. doi: 10.1016/j.mcpro.2024.100804. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

