IECata: interpretable bilinear attention network and evidential deep learning improve the catalytic efficiency prediction of enzymes
- PMID: 40548541
- PMCID: PMC12205960
- DOI: 10.1093/bib/bbaf283
IECata: interpretable bilinear attention network and evidential deep learning improve the catalytic efficiency prediction of enzymes
Abstract
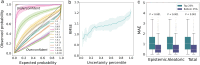
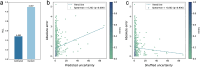
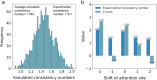
Enzyme catalytic efficiency (kcat/Km) is a key parameter for identifying high-activity enzymes. Recently, deep learning techniques have demonstrated the potential for fast and accurate kcat/Km prediction. However, three challenges remain: (i) the limited size of the available kcat/Km dataset hinders the development of deep learning models; (ii) the model predictions lack reliable confidence estimates; and (iii) models lack interpretable insights into enzyme-catalyzed reactions. To address these challenges, we proposed IECata, a kcat/Km prediction model that provides uncertainty estimation and interpretability. IECata collected a dataset of 11 815 kcat/Km entries from the BRENDA and SABIO-RK databases, along with an out-of-domain test dataset of 806 entries from the literature. By introducing evidential deep learning, IECata provides uncertainty estimates for kcat/Km predictions. Moreover, it uses a bilinear attention mechanism to focus on learning crucial local interactions to interpret the key residues and substrate atoms in enzyme-catalyzed reactions. Testing results indicate that the prediction performance of IECata exceeds that of state-of-the-art benchmark models. More importantly, it provides a reliable confidence assessment for these predictions. Case studies further highlight that the incorporation of uncertainty in screening for highly active enzymes can effectively increase the hit ratio, thereby improving the efficiency of experimental validation and accelerating directed enzyme evolution. To facilitate researchers' use of IECata, we have developed an online prediction platform: http://mathtc.nscc-tj.cn/cataai/.
Keywords: k cat/Km prediction; bilinear attention mechanism; evidential deep learning; interpretability; uncertainty.
© The Author(s) 2025. Published by Oxford University Press.
Figures






References
-
- Chronopoulou EG, Labrou NE. Site-saturation mutagenesis: a powerful tool for structure-based design of combinatorial mutation libraries. Curr Protoc Protein Sci 2011;26:26.6.1–10. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

