Synthetic host defense peptide inhibits SARS-CoV-2 replication in vitro
- PMID: 40548715
- PMCID: PMC12326988
- DOI: 10.1128/aac.01700-24
Synthetic host defense peptide inhibits SARS-CoV-2 replication in vitro
Abstract
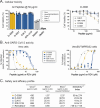
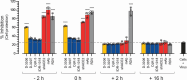
Although myriads of potential antiviral agents have been tested against SARS-CoV-2, only a handful have proven to be effective in clinical trials. During the COVID-19 pandemic, many known or novel peptides were evaluated for their ability to inhibit SARS-CoV-2 replication; however, testing of D-enantiomers that resist body and viral proteases has been limited. Here, we characterized the ability of D-3006, a D-enantiomeric synthetic host defense peptide, to inhibit SARS-CoV-2 replication in vitro. A battery of authentic SARS-CoV-2 variants (ancestral, Mu, Delta, and Omicron BA.1) and a comprehensive panel of β-coronavirus spike pseudotyped lentiviruses were used to demonstrate that D-3006 safely (CC50value = 430 µg/mL) blocked spike-mediated entry (EC50 values ranging from 1.57 to 5.37 µg/mL) and also had synergistic anti-SARS-CoV-2 activity in vitro when combined with the viral polymerase inhibitor remdesivir. We also showed that D-3006 inhibited influenza A virus (H1N1) replication in vitro, suggesting that this synthetic host defense peptide could have potential broad antiviral activity against multiple enveloped viruses. These data, together with negative-stain transmission electron microscopy analysis, suggest that the mechanism of action of D-3006 is associated with non-specific binding to the viral membrane, most likely causing virus aggregation and interfering with virus attachment and entry. The potential broad-spectrum antiviral activity of D-3006, its innate resistance to host proteases, as well as the possibility of being used in combination with other antiviral drugs suggest that this host synthetic peptide could be developed as a candidate for the treatment of SARS-CoV-2 and/or other respiratory viral infections.
Keywords: COVID-19; SARS-CoV-2; antiviral; host defense peptides; influenza virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
Targeting SARS-CoV-2 RNA-dependent RNA polymerase with the coumarin derivative BPR2-D2: Evidence from cell-based and enzymatic studies.Biomed Pharmacother. 2025 Aug;189:118252. doi: 10.1016/j.biopha.2025.118252. Epub 2025 Jun 20. Biomed Pharmacother. 2025. PMID: 40543163
-
Determinants of susceptibility to SARS-CoV-2 infection in murine ACE2.J Virol. 2025 Jun 17;99(6):e0054325. doi: 10.1128/jvi.00543-25. Epub 2025 May 12. J Virol. 2025. PMID: 40353671 Free PMC article.
-
Different SARS-CoV-2 variants inhibited by RRM designed peptide.PLoS One. 2025 Jul 22;20(7):e0327582. doi: 10.1371/journal.pone.0327582. eCollection 2025. PLoS One. 2025. PMID: 40694559 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
-
Measures implemented in the school setting to contain the COVID-19 pandemic.Cochrane Database Syst Rev. 2022 Jan 17;1(1):CD015029. doi: 10.1002/14651858.CD015029. Cochrane Database Syst Rev. 2022. Update in: Cochrane Database Syst Rev. 2024 May 2;5:CD015029. doi: 10.1002/14651858.CD015029.pub2. PMID: 35037252 Free PMC article. Updated.
References
-
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017 - DOI - PMC - PubMed
-
- Wu N, Joyal-Desmarais K, Ribeiro PAB, Vieira AM, Stojanovic J, Sanuade C, Yip D, Bacon SL. 2023. Long-term effectiveness of COVID-19 vaccines against infections, hospitalisations, and mortality in adults: findings from a rapid living systematic evidence synthesis and meta-analysis up to December, 2022. Lancet Respir Med 11:439–452. doi: 10.1016/S2213-2600(23)00015-2 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

