Isolation, identification, and pathogenicity of Pseudoalteromonas aliena associated with oyster disease outbreaks in summer
- PMID: 40548722
- PMCID: PMC12323356
- DOI: 10.1128/spectrum.00173-25
Isolation, identification, and pathogenicity of Pseudoalteromonas aliena associated with oyster disease outbreaks in summer
Abstract
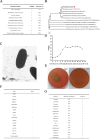
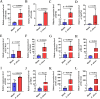
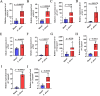
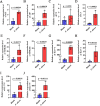
With the continuous expansion of oyster farming scale, disease has become one of the main obstacles to restricting the development of oyster farming. In the present study, 20 bacterial strains were identified from Crassostrea gigas with pustulosis, among which Pseudoalteromonas aliena emerged as the predominant strain, characterized by its rod-shaped morphology and possession of flagella. P. aliena exhibited α-hemolytic activity at 28°C and displayed high susceptibility to all 20 chemotherapeutic agents tested. After P. aliena infection, the oyster mortality rate increased. The gills were swollen and eroded, and the mantle was green with pustules after P. aliena infection. The gill filaments exhibited swelling and necrotic cells, and the mantle showed a loose histological structure with cavities and disruption of epithelial cells. The extracellular products (ECPs) from P. aliena had urease, protease, and amylase activities. The potential virulence proteins identified from ECPs were GroL, ClpB, and HtpG proteins. After injection with ECPs, there was an increase in the oyster mortality rate, and the observed symptoms in gill filaments and mantle were consistent with those observed after P. aliena infection. In addition, the mRNA expressions of inflammation- and programmed cell death-related genes were significantly upregulated in gills and mantle. The relative abundances of Vibrio, Arcobacter, and Pseudoalteromonas also exhibited a significant increase in the gills and mantle. The results demonstrated that P. aliena was the pathogenic bacterium for oysters, and its pathogenicity mechanism was systematically clarified, which provided valuable insights for the prevention and control of bacterial disease in oysters.IMPORTANCEDisease has currently emerged as one of the principal impediments to restricting the development of the oyster breeding industry. In the present study, Pseudoalteromonas aliena was identified from Crassostrea gigas with pustulosis. After P. aliena infection, the oyster mortality rate increased. The gills were swollen and eroded, and the mantle was green with pustules. Extracellular products (ECPs) from P. aliena had urease, protease, and amylase activities. The potential virulence proteins identified from ECPs were GroL, ClpB, and HtpG proteins. After injection with ECPs, the oyster mortality rate increased. The mRNA expressions of inflammation- and programmed cell death-related genes in gills and mantle increased significantly, and the relative abundances of Vibrio, Arcobacter, and Pseudoalteromonas exhibited a significant increase after P. aliena infection. The results demonstrated that P. aliena was the pathogenic bacterium for oysters, and its pathogenicity mechanism was systematically clarified, which provided valuable insights for the prevention and control of bacterial disease in oysters.
Keywords: 16S rRNA; Crassostrea gigas; Pseudoalteromonas aliena; inflammation; pathogenicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Gómez-León J, Villamil L, Lemos ML, Novoa B, Figueras A. 2005. Isolation of Vibrio alginolyticus and Vibrio splendidus from aquacultured carpet shell clam (Ruditapes decussatus) larvae associated with mass mortalities. Appl Environ Microbiol 71:98–104. doi: 10.1128/AEM.71.1.98-104.2005 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 32222086,32230110/National Natural Science Foundation of China
- CARS-49/the fund for China Agriculture Research System
- the fund for Outstanding Talents and Innovation Teams of Agricultureal Scientific Research in MARA
- LT202009/the innovation team of Agriculture Environment Safety from Liaoning Province
- XLYC2203087/Liaoning Revitalization Talents Program
LinkOut - more resources
Full Text Sources

