Circadian Gene NPAS2 Relieves Hypertrophic Scar Formation via CDC25A-Mediated Fibroblasts Activity
- PMID: 40548841
- PMCID: PMC12184073
- DOI: 10.1111/jcmm.70643
Circadian Gene NPAS2 Relieves Hypertrophic Scar Formation via CDC25A-Mediated Fibroblasts Activity
Abstract
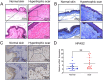
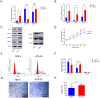
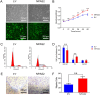
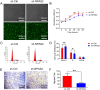
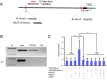
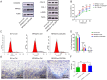
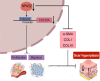
Neuronal PAS domain protein 2 (NPAS2) is critical in tissue fibrosis. Hypertrophic scars (HTS), a form of skin fibrosis, are characterised by excessive myofibroblast proliferation and abnormal extracellular matrix (ECM) deposition. However, whether NPAS2 contributes to skin fibrosis and the development of HTS remains unclear. In this study, the expression of NPAS2 between normal skin and hypertrophic scars (HTS) was assessed using RT-qPCR and immunohistochemistry (IHC). Human dermal fibroblasts (HDFs) and HTS-derived fibroblasts (HTS-Fs) were isolated from normal skin and HTS, respectively. NPAS2 was knocked down in HTS-Fs and overexpressed in HDFs via gene transfection. Cell proliferation and migration of transfected HTS-Fs and HDFs were analysed using flow cytometry, CCK-8 and transwell assays. The expressions of NPAS2, CLOCK, BMAL1, COL I, COL III, α-SMA and CDC25A were evaluated by western blotting and RT-qPCR. Dual-luciferase reporter assays and chromatin immunoprecipitation (ChIP) identified the regulatory effect of NPAS2 on CDC25A. In vivo, an 8 × 8 mm full-thickness skin defect was created on the tail of SD rats, with viral particles (1 × 107) of r-plenR-sh-NPAS2 or r-plenR-NPAS2-NC injected subcutaneously at the wound edges weekly. Tissue samples, histopathological analyses and photographs were taken until the wound healed completely. The results indicated that NPAS2 was significantly upregulated in HTS. The proliferation, migration, and expression of COL I, COL III, and α-SMA were higher in HDFs overexpressing NPAS2 than those of HDFs themselves. In contrast, the behaviours mentioned above of HTS-Fs knocking down NPAS2 were lower than that of HTS-Fs. Mechanistically, the migration and proliferation promoting effect of NPAS2 was mediated by the binding of NPAS2 to the E-like-box of CDC25A. In vivo, compared with the r-plenR-NPAS2-NC group, the re-epithelialised regions of r-plenR-sh-NPAS2 were pink, flat and as large as the initial wound. In addition, their dermal structures were similar to skin and possessed loose and regular collagen arrangement which was parallel to the epidermis. Take together, these findings suggested that compared with HDFs, NPAS2 was upregulated in HTS-Fs. NPAS2 promoted the activation of HDFs, which is characterised by stronger proliferation and migration and the higher level of α-SMA, COL I and COL III. In which, the proliferation and migration effects of NPAS2 were mediated by CDC25A. Furthermore, NPAS2 knocked down in rat tail wounds inhibited the HTS formation. Therefore, NPAS2 may serve as a potential therapeutic target for HTS in the future.
Keywords: CDC25A; NPAS2; circadian clock; fibrosis; hypertrophic scar.
© 2025 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- John W. L., Shawn T. M., Katherine S., and Matthew B. K., “Epidemiology and Impact of Scarring After Burn Injury: A Systematic Review of the Literature,” Journal of Burn Care & Research 33, no. 1 (2011): 136–146. - PubMed
-
- Peter O. K. and Edward E. T., “Biological Principles of Scar and Contracture,” Hand Clinics 33, no. 2 (2017): 277–292. - PubMed
-
- Frech F. S., Hernandez L., Urbonas R., Zaken G. A., Dreyfuss I., and Nouri K., “Hypertrophic Scars and Keloids: Advances in Treatment and Review of Established Therapies,” American Journal of Clinical Dermatology 24, no. 2 (2023): 225–245. - PubMed
MeSH terms
Substances
Grants and funding
- 2020Y9094/Joint Funds for the innovation of science and Technology, Fujian province
- 2023J01692/Fujian Provincial Natural Science Foundation of China
- ([2021]76)/The High-level Hospital and Clinical Specialty Discipline Construction Program for Fujian Medical Development, China
- Fujian Provincial Key Laboratory of Burn and Trauma, China
LinkOut - more resources
Full Text Sources

