The Non-Canonical ChREBPα Activity Suppresses the Activation of Hepatic Stellate Cells and Liver Fibrosis by Antagonizing TGF-β-E2F1 Axis
- PMID: 40548862
- PMCID: PMC12362833
- DOI: 10.1002/advs.202415032
The Non-Canonical ChREBPα Activity Suppresses the Activation of Hepatic Stellate Cells and Liver Fibrosis by Antagonizing TGF-β-E2F1 Axis
Abstract
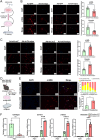
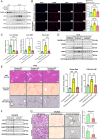
Sustained activation of hepatic stellate cells (HSCs) drives liver fibrosis in response to chronic liver injury and inflammation. It is reported that profibrogenic signals released from stressed/injured hepatocytes evoke fibrogenic responses in HSCs. However, intrahepatocyte players that modulate such cell-to-cell communications remain poorly defined. In this study, hepatic ChREBPα is found to be reduced in mouse models of chemical-induced liver fibrosis as well as in three groups of human patients with liver fibrosis. Chrebpα-LKO mice are highly sensitive to both chemical (CCL4 and TAA) and bile duct ligation (BDL)-induced liver injury and developed more advanced liver fibrosis without affecting liver lipid content. Hepatocyte ChREBPα overexpression suppressed the activation of HSCs in an in vitro medium transfer experiment in part via inhibiting the expression of profibrogenic factors THBS1 and CTGF. RNA-Seq analysis revealed that E2F1, a novel effector of TGFβ-mediated fibrogenic pathway, is highly induced in the liver of Chrebpα-LKO mice. Hepatic knockdown of E2F1 ameliorated the increased liver fibrosis in mice with hepatic Chrebpα deficiency while reducing the expression of hepatic THBS1 and CTGF.
Keywords: ChREBPα; E2F1; TGF‐β signaling; hepatic stellate cells; liver fibrosis.
© 2025 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Zamani M., Alizadeh‐Tabari S., Ajmera V., Singh S., Murad M. H., Loomba R., Clin. Gastroenterol. Hepatol. 2024, 23, 1123. - PubMed
-
- a) Lee Y. S., Seki E., Cell Mol. Gastroenterol. Hepatol. 2023, 16, 355; - PMC - PubMed
- b) Seki E., Brenner D. A., J. Hepatobiliary Pancreat. Sci. 2015, 22, 512; - PMC - PubMed
- c) Hammerich L., Tacke F., Nat. Rev. Gastroenterol. Hepatol. 2023, 20, 633; - PubMed
- d) Dhar D., Baglieri J., Kisseleva T., Brenner D. A., Exp. Biol. Med. (Maywood) 2020, 245, 96; - PMC - PubMed
- e) Kim H. Y., Sakane S., Eguileor A., Carvalho Gontijo Weber R., Lee W., Liu X., Lam K., Ishizuka K., Rosenthal S. B., Diggle K., Brenner D. A., Kisseleva T., Cell Mol. Gastroenterol. Hepatol. 2024, 17, 93. - PMC - PubMed
-
- a) Tsuchida T., Friedman S. L., Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397; - PubMed
- b) Kisseleva T., Brenner D., Nat. Rev. Gastroenterol. Hepatol. 2020, 18, 151; - PubMed
- c) Liu X., Xu J., Rosenthal S., Zhang L. J., McCubbin R., Meshgin N., Shang L., Koyama Y., Ma H. Y., Sharma S., Heinz S., Glass C. K., Benner C., Brenner D. A., Kisseleva T., Gastroenterology 2020, 158, 1728. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
