In vitro programmable DNA cleavage by a eukaryotic Argonaute
- PMID: 40548933
- PMCID: PMC12205983
- DOI: 10.1093/nar/gkaf561
In vitro programmable DNA cleavage by a eukaryotic Argonaute
Erratum in
-
Correction to 'In vitro programmable DNA cleavage by a eukaryotic Argonaute'.Nucleic Acids Res. 2025 Jul 8;53(13):gkaf690. doi: 10.1093/nar/gkaf690. Nucleic Acids Res. 2025. PMID: 40671530 Free PMC article. No abstract available.
Abstract
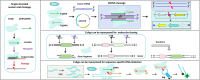
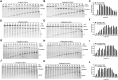
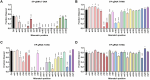
Eukaryotic Argonautes (eAgos) have traditionally been characterized by their ability to utilize RNA guides to identify RNA targets, thereby engaging in post-transcriptional gene silencing pathways. While some eAgos have been demonstrated to use DNA guides for RNA cleavage, the ability of eAgos to cleave DNA targets remains unclear. In this study, we characterized CsAgo, an eAgo protein derived from thermophilic eukaryote Chaetomiumsp. MPI-CAGE-AT-0009, demonstrating a novel ability to cleave both DNA and RNA targets in vitro. Guided by short single-stranded DNA (ssDNA) or RNA, CsAgo exhibits robust RNA cleavage activity at 20-90°C in vitro. CsAgo can effectively cleave ssDNA guided by RNA guides at 20-50°C in vitro. Notably, CsAgo can utilize DNA guides to effectively cleave ssDNA, plasmid double-stranded DNA (dsDNA), and linear dsDNA at ≥80°C in vitro. Based on its ability to cleave dsDNA at high temperatures, CsAgo demonstrates versatility and efficacy in simplifying routine cloning workflows. Additionally, we have developed a CsAgo-based nucleic acid detection method based on a Pyrococcus furiosus Ago-mediated nucleic acid detection method, which exhibits a high sensitivity of six copies/reaction. These results suggest that eAgos include that, in theory, can be utilized for potential DNA-targeting applications. This not only enhances our understanding of eAgos but also expands the toolkit for DNA manipulation.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
L.M., F.W., G.Y., X.Y., W.L., Y.L., and W.C. are co-inventors on a patent application (202410427777.X) related to this work filed by Hubei University.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

