Systematic analysis of siRNA and mRNA features impacting fully chemically modified siRNA efficacy
- PMID: 40548938
- PMCID: PMC12205987
- DOI: 10.1093/nar/gkaf479
Systematic analysis of siRNA and mRNA features impacting fully chemically modified siRNA efficacy
Abstract
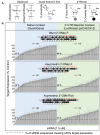
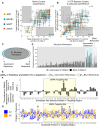
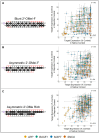
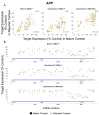
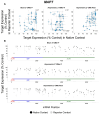
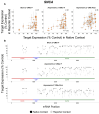
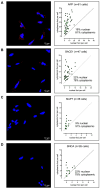
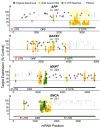
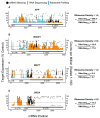
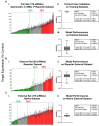
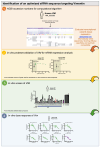
Chemically modified small interfering RNAs (siRNAs) are a promising drug class that silences disease-causing genes via mRNA degradation. Both siRNA-specific features (e.g. sequence, modification pattern, and structure) and target mRNA-specific factors contribute to observed efficacy. Systematically defining the relative contributions of siRNA sequence, structure, and modification pattern versus the native context of the target mRNA is necessary to inform design considerations and facilitate the widespread application of this therapeutic platform. To address this, we synthesized a panel of ∼1260 differentially modified siRNAs and evaluated their silencing efficiency against therapeutically relevant mRNAs (APP, BACE1, MAPT, and SNCA) using both reporter-based and native expression assays. Our results demonstrate that the siRNA modification pattern (e.g. level of 2'-O-methyl content) significantly impacts efficacy, while structural features (e.g. symmetric versus asymmetric configurations) do not. Furthermore, we observed substantial differences in the number of effective siRNAs identified per target. These target-specific differences in hit rates are largely mitigated when efficacy is tested in the context of a reporter assay, confirming that native mRNA-specific features influence siRNA performance. Key target-specific factors, including exon usage, polyadenylation site selection, and ribosomal occupancy, partially explained efficacy variability. These insights led to a proposed framework of parameters for optimizing therapeutic siRNA design.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
A.K. is an inventor on Fully Stabilized Asymmetric siRNA, U.S. Utility App. No. 15/089,423; A.K. and S.M.D. are inventors on O-Methyl Rich Fully Stabilized Oligonucleotides, U.S. Utility App. No. 16/550,076; A.K., S.M.D., and C.F. are inventors on Oligonucleotides for APP Modulation. U.S. Utility App. No. 18/120,030; A.K., S.M.D., K.M., and C.F. are inventors on Oligonucleotides for MAPT Modulation. U.S. Utility App. No. 17/204,480; A.K., S.M.D., K.M., and C.F. are inventors on Oligonucleotides for SNCA Modulation. U.S. Utility App. No. 17/204,483; A.K., S.M.D., S.H., K.M., J.S., N.H., C.F., and V.N.H. are inventors on Development of lung-active siRNAs and ASOs for the prophylaxis and treatment of COVID-19 Infection. U.S. Utility App. No. 17/333,839.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

