USP37 counteracts HLTF to protect damaged replication forks and promote survival of BRCA1-deficient cells and PARP inhibitor resistance
- PMID: 40548939
- PMCID: PMC12205991
- DOI: 10.1093/nar/gkaf544
USP37 counteracts HLTF to protect damaged replication forks and promote survival of BRCA1-deficient cells and PARP inhibitor resistance
Abstract
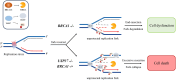
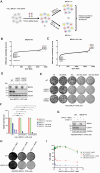
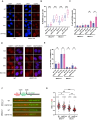
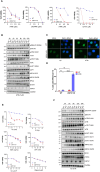
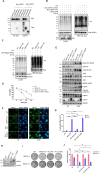
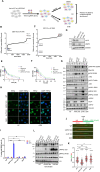
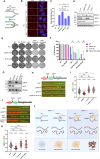
Poly(ADP-ribose) polymerase inhibitors (PARPi) have greatly improved survival of cancer patients harboring BRCA1 mutations. However, therapy resistance develops via either restoration of homologous recombination or replication fork stabilization. Therapeutic targets to overcome PARPi resistance are critically needed. We identified the deubiquitinase USP37 as a key determinant of PARPi toxicity in BRCA1-deficient cells via whole-genome CRISPR screens. USP37 ablation enhanced PARPi sensitivity in BRCA1-deficient cells and also overcame PARPi resistance due to 53BP1 loss. USP37 interacts with and deubiquitinates replication protein A (RPA) at stalled replication forks to limit excessive RPA accumulation, progressive RPA exhaustion, and the conversion of RPA-coated single-stranded DNAs to DNA double-strand breaks. Moreover, USP37 limits helicase-like transcription factor (HLTF) accumulation at replication forks and thus prevents MRE11-dependent fork degradation upon replication stress. Depletion of HLTF reversed the replication-associated damage observed in USP37 knockout cells. Our data suggest that USP37 protects replication fork stability by counteracting HLTF function and promotes survival of BRCA1-deficient cells, making it a promising drug target to overcome PARPi resistance in BRCA1-deficient tumors.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures
References
MeSH terms
Substances
Grants and funding
- R01 CA210929/CA/NCI NIH HHS/United States
- 82403439/National Natural Science Foundation of China
- CA216437/GF/NIH HHS/United States
- Jiangsu Specially Appointed Professorship Foundation
- CA193124/GF/NIH HHS/United States
- BK20241138/Natural Science Foundation of Jiangsu Province of China
- CA216911/GF/NIH HHS/United States
- RP180813/CPRIT
- RP160667/CPRIT
- CA210929/GF/NIH HHS/United States
- R01 CA216437/CA/NCI NIH HHS/United States
- R01 CA216911/CA/NCI NIH HHS/United States
- 82404048/National Natural Science Foundation of China
- R01 CA275712/CA/NCI NIH HHS/United States
- CA275712/GF/NIH HHS/United States
- CA274234/GF/NIH HHS/United States
- R35 CA274234/CA/NCI NIH HHS/United States
- BK20240730/Natural Science Foundation of Jiangsu Province of China
- P01 CA193124/CA/NCI NIH HHS/United States
- Pamela and Wayne Garrison Distinguished Chair in Cancer Research
LinkOut - more resources
Full Text Sources
Miscellaneous

