What makes a small RNA work?
- PMID: 40548940
- PMCID: PMC12205990
- DOI: 10.1093/nar/gkaf563
What makes a small RNA work?
Abstract
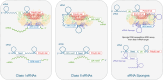
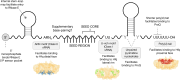
Bacterial small RNAs (sRNAs) are key regulators of gene expression, interacting with target messenger RNAs (mRNAs) through imperfect base pairing. Unlike other non-coding RNAs such as microRNAs and PIWI-interacting RNAs, bacterial sRNAs exhibit significant sequence and structural diversity, complicating functional predictions. Recent high-throughput profiling of the sRNA interactome has accentuated this problem by revealing a highly complex network of sRNA interactions. It is clear that there is an incredible diversity of sRNA interactions with different RNA classes in vivo, including different interaction modes with mRNAs. In this review, we attempt to summarize the known sequence and structural features that contribute to sRNA function in bacteria. As many of these features drive recruitment of protein partners, we necessarily focus on interactions with chaperones and ribonucleases, the best studied being Hfq and RNase E. Where possible, we have included examples outside this well-studied system as diversity and rule breaking appear to be central themes of sRNA biology. Understanding the sequences and structures that drive sRNA function will enhance our ability to predict regulatory outcomes, and this may inform the development of effective RNA therapeutics that are inspired by bacterial sRNA mechanisms.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

