Building a neural network model to define DNA sequence specificity in V(D)J recombination
- PMID: 40548941
- PMCID: PMC12205992
- DOI: 10.1093/nar/gkaf551
Building a neural network model to define DNA sequence specificity in V(D)J recombination
Abstract
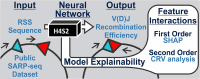
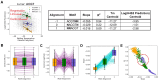
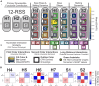
In developing lymphocytes, V(D)J recombination assembles functional antigen receptor (AgR) genes through rearrangement of the AgR loci to adjoin component gene segments. Each candidate gene segment for recombination is flanked by a recombination signal sequence (RSS), composed of heptamer and nonamer motifs separated by 12 or 23 base pairs. To initiate V(D)J recombination, the recombination activating proteins RAG1 and RAG2 create DNA double-stranded breaks between a 12/23-RSS pair and their adjoining gene segments. The basis for selection of individual RSSs during each V(D)J recombination event is not well understood due, in part, to the wide-spread distribution of the semi-conserved RSSs across the AgR loci. Using publicly-available data for V(D)J recombination efficiencies on randomized 12-RSSs, we first built a neural network model that delineates how changes in sequence at certain positions in the RSS affects recombination efficiency. Second, to interpret the model's decision-making process, we repurposed the game theoretic SHapley Additive exPlanations (SHAP) approach, with the results illustrating how nucleotides at pairwise positions in the heptamer provide synergistic contributions to recombination efficiency. Third, we trained a nonamer-informed neural network model with varied nonamer RSS substrates, and subsequently identified interdependent effects between the heptamer and nonamer regions on recombination efficiency.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures







References
-
- Schatz DG, Zhang Y, Xiao J et al.. Honjo T, Reth M, Radbruch A, Alt F, Martin A Molecular Biology of B Cells. 2024; 3rd editionCambridge, Massachusetts, United States: Academic Press; 13–57.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

