Novel drug-inducible CRISPRa/i systems for rapid and reversible manipulation of gene transcription
- PMID: 40549156
- PMCID: PMC12185803
- DOI: 10.1007/s00018-025-05786-7
Novel drug-inducible CRISPRa/i systems for rapid and reversible manipulation of gene transcription
Erratum in
-
Correction: Novel drug-inducible CRISPRa/i systems for rapid and reversible manipulation of gene transcription.Cell Mol Life Sci. 2025 Nov 14;82(1):407. doi: 10.1007/s00018-025-05914-3. Cell Mol Life Sci. 2025. PMID: 41236543 Free PMC article. No abstract available.
Abstract
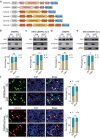
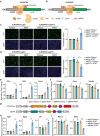
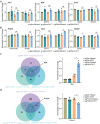
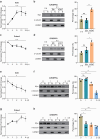
CRISPR activation and interference (CRISPRa/i) are highly effective tools to regulate transcription by fusing dead Cas9 (dCas9) with transcriptional regulatory factors guided by small guide RNA (sgRNA) in mammalian cells and mice. Still, a controllable gene regulation system is desired to investigate and manipulate dynamic biological processes. Here, we reported flexible drug-responsive CRISPRa/i systems by fusing mutated human estrogen receptor (ERT2) domains, which responded to estrogen analogue tamoxifen or its active metabolite 4-hydroxy-tamoxifen (4OHT), to CRISPRa/i components for transcriptional regulation. Upon 4OHT treatment, the optimal variants, ERT2-ERT2-CRISPRa/i-ERT2 (iCRISPRa/i), showed rapid protein translocation of iCRISPRa/i from cytoplasm to nucleus and subsequent transcriptional response. The inducible transcriptional manipulation could be restored to its original level when 4OHT was withdrawn. Moreover, the efficiencies of gene expression regulation of iCRISPRa/i were comparable to those of non-inducible and doxycycline-inducible counterparts, with a lower leakage and a faster drug response activity. The iCRISPRa/i systems successfully induced phenotypic changes in various cell lines. These results highlight that iCRISPRa/i systems could achieve fast and flexible drug-responsive transcriptional modulation and phenotypic changes, and thus provide better options for gain- and loss-of-function model construction and gene therapy.
Keywords: 4OHT; Phenotypic change; Transcriptional regulation; iCRISPRa/i.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Conflict of interest: The authors declare no conflict of interest. Ethical standards: The experiments comply with the current laws of the country where we were performed. Consent to publish: All the authors have read and approved the final version of the manuscript and agreed to its publication in Cellular and Molecular Life Sciences.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

