JunB-HBZ nuclear translocation by TGF-β is a key driver in HTLV-1-mediated leukemogenesis
- PMID: 40549917
- PMCID: PMC12232710
- DOI: 10.1073/pnas.2420756122
JunB-HBZ nuclear translocation by TGF-β is a key driver in HTLV-1-mediated leukemogenesis
Abstract
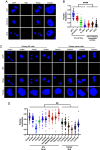
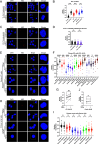
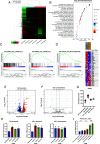
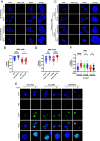
The HTLV-1 bZIP factor (HBZ) gene, which is the only viral gene conserved and consistently expressed in all adult T-cell leukemia-lymphoma (ATL) cases, is critical for ATL oncogenesis. Although HBZ protein is found in both the nucleus and the cytoplasm, the dynamics of HBZ protein localization and its contribution to oncogenesis have not been fully elucidated. In this study, we analyzed the subcellular expression pattern of HBZ in primary HTLV-1-infected T cells from asymptomatic carriers and leukemic cells of ATL patients using the Proximity Ligation Assay. Nuclear localization of HBZ protein was significantly higher in fresh ATL cells than in HTLV-1-infected cells from carriers. Importantly, translocation of HBZ protein from the cytoplasm to the nucleus after TGF-β activation was observed in ATL patients, but not in HTLV-1 carriers. In ATL cells, the cellular transcription factors JunB and pSmad3 interact with HBZ and facilitate its nuclear translocation upon TGF-β stimulation. JUNB knockdown inhibits cell proliferation in vitro and in vivo and promotes apoptosis in ATL cells but not in HTLV-1-infected nonleukemic cells, indicating that JunB has important roles in maintaining ATL cells. In conclusion, TGF-β-induced nuclear translocation of HBZ-JunB complexes is associated with ATL oncogenesis.
Keywords: ATL; HBZ; HTLV-1; JunB; TGF-β/Smad signaling pathway.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

Similar articles
-
HTLV-I basic leucine zipper factor gene mRNA supports proliferation of adult T cell leukemia cells.Proc Natl Acad Sci U S A. 2006 Jan 17;103(3):720-5. doi: 10.1073/pnas.0507631103. Epub 2006 Jan 9. Proc Natl Acad Sci U S A. 2006. PMID: 16407133 Free PMC article.
-
The Role of HBZ in HTLV-1-Induced Oncogenesis.Viruses. 2016 Feb 2;8(2):34. doi: 10.3390/v8020034. Viruses. 2016. PMID: 26848677 Free PMC article. Review.
-
Localization, quantification and interaction with host factors of endogenous HTLV-1 HBZ protein in infected cells and ATL.Retrovirology. 2015 Jul 4;12:59. doi: 10.1186/s12977-015-0186-0. Retrovirology. 2015. PMID: 26140924 Free PMC article.
-
HTLV-I Basic Leucine Zipper Factor (sHBZ) Actively Associates with Nucleophosmin (B23) in the Nucleolus.Viruses. 2025 May 19;17(5):727. doi: 10.3390/v17050727. Viruses. 2025. PMID: 40431738 Free PMC article.
-
Adult T-cell leukemia-lymphoma as a viral disease: Subtypes based on viral aspects.Cancer Sci. 2021 May;112(5):1688-1694. doi: 10.1111/cas.14869. Epub 2021 Mar 30. Cancer Sci. 2021. PMID: 33630351 Free PMC article. Review.
References
-
- Heldin C. H., Miyazono K., ten Dijke P., TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature 390, 465–471 (1997). - PubMed
-
- Proietti F. A., Carneiro-Proietti A. B., Catalan-Soares B. C., Murphy E. L., Global epidemiology of HTLV-I infection and associated diseases. Oncogene 24, 6058–6068 (2005). - PubMed
-
- Matsuoka M., Jeang K. T., Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat. Rev. Cancer 7, 270–280 (2007). - PubMed
MeSH terms
Substances
Grants and funding
- 20cm0106306h0005/Japan Agency for Medical Research and Development (AMED)
- 20fk0108088h0002/Japan Agency for Medical Research and Development (AMED)
- 21am0401003h0003/Japan Agency for Medical Research and Development (AMED)
- 19H03689/MEXT | Japan Society for the Promotion of Science (JSPS)
- 23H02936/MEXT | Japan Society for the Promotion of Science (JSPS)
- 23K15303/MEXT | Japan Society for the Promotion of Science (JSPS)
- 24fk0108629h0003/Japan Agency for Medical Research and Development (AMED)
- 25K11650/MEXT | Japan Society for the Promotion of Science (JSPS)
- 24K22136/MEXT | Japan Society for the Promotion of Science (JSPS)
- 25K19580/MEXT | Japan Society for the Promotion of Science (JSPS)
LinkOut - more resources
Full Text Sources
Miscellaneous

