Developing a new cleavable crosslinker reagent for in-cell crosslinking
- PMID: 40550823
- PMCID: PMC12185727
- DOI: 10.1038/s42004-025-01568-1
Developing a new cleavable crosslinker reagent for in-cell crosslinking
Abstract
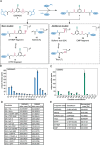
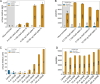
Crosslinking mass spectrometry (XL-MS) is a powerful technology that recently emerged as an essential complementary tool for elucidating protein structures and mapping interactions within a protein network. Crosslinkers which are amenable to post-linking backbone cleavage simplify peptide identification, aid in 3D structure determination and enable system-wide studies of protein-protein interactions (PPIs) in cellular environments. However, state-of-the-art cleavable linkers are fraught with practical limitations, including extensive evaluation of fragmentation energies and fragmentation behavior of the crosslinker backbone. We herein introduce DiSPASO (bis(2,5-dioxopyrrolidin-1-yl) 3,3'-((5-ethynyl-1,3-phenylene)bis(methylenesulfinyl))dipropanoate) as a lysine-selective, MS-cleavable crosslinker with an alkyne handle for affinity enrichment. DiSPASO was designed and developed for efficient cell membrane permeability and crosslinking while securing low cellular perturbation. We tested DiSPASO employing three different copper-based enrichment strategies using model systems with increasing complexity (Cas9-Halo, purified ribosomes, live cells). Fluorescence microscopy in-cell crosslinking experiments revealed a rapid uptake of DiSPASO into HEK 293 cells within 5 minutes. While DiSPASO represents progress in cellular PPI analysis, its limitations and low crosslinking yield in cellular environments require careful optimisation of the crosslinker design, highlighting the complexity of developing effective XL-MS tools and the importance of continuous innovation in accurately mapping PPI networks within dynamic cellular environments.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
LinkOut - more resources
Full Text Sources
Miscellaneous

