Predicting resistance to chemotherapy using chromosomal instability signatures
- PMID: 40551015
- PMCID: PMC12283407
- DOI: 10.1038/s41588-025-02233-y
Predicting resistance to chemotherapy using chromosomal instability signatures
Abstract
Chemotherapies are often given without precision biomarkers, exposing patients to toxic side effects without guaranteed benefit. Here we present chromosomal instability signature biomarkers that identify resistance to platinum-, taxane- and anthracycline-based treatments using a single genomic test. In retrospectively emulated randomized-control biomarker clinical trials using real-world cohorts (n = 840), predicted resistant patients had elevated treatment failure risk for taxane (hazard ratio (HR) of 7.44) and anthracycline (HR of 1.88) in ovarian, taxane (HR of 3.98) and anthracycline (HR of 3.69) in metastatic breast and taxane (HR of 5.46) in metastatic prostate. Nonrandomized emulations showed predictive capacity for platinum resistance in ovarian (HR of 1.46) and anthracycline in sarcoma (HR of 3.59). We demonstrate feasibility using whole-genome sequencing, capture-panel sequencing and cell-free DNA. Our findings highlight the clinical value of chromosomal instability signatures in predicting resistance to chemotherapies across multiple cancer types, with the potential to transform the one-size-fits-all chemotherapy approach into precise, tailored treatment.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: G.M., A.M.P., J.D.B., J.Y. and F.M. are co-founders, directors and shareholders of Tailor Bio Ltd. J.S.T., L.M., D.G.-L., A.R., O.A. and A.E.C. are current or recent employees and shareholders of Tailor Bio Ltd. M.E.-R., W.-K.L., H.D. and D.D.S. are current or previous employees of Tailor Bio Ltd. G.M., F.M., A.M.P. and J.D.B. are inventors on a patent on using copy number signatures to predict response to doxorubicin treatment in ovarian cancer (patent no. PCT/EP2021/065058). G.M., B.H. and F.M. are inventors on a patent on a method for identifying pan-cancer copy number signatures (patent no. PCT/EP2022/077473). G.M., D.G.-L. and D.G.-S. are inventors on a patent on a method for capture bias correction for copy number calling in targeted sequencing data (patent no. PCT/EP2024/070580). The other authors declare no competing interests.
Figures
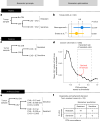

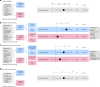

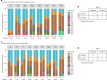



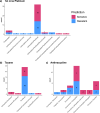


References
-
- Kuderer, N. M., Desai, A., Lustberg, M. B. & Lyman, G. H. Mitigating acute chemotherapy-associated adverse events in patients with cancer. Nat. Rev. Clin. Oncol.19, 681–697 (2022). - PubMed
-
- Jørgensen, J. T. Companion and complementary diagnostics: clinical and regulatory perspectives. Trends Cancer2, 706–712 (2016). - PubMed
-
- Hunter, D. J., Khoury, M. J. & Drazen, J. M. Letting the genome out of the bottle—will we get our wish? N. Engl. J. Med.358, 105–107 (2008). - PubMed
-
- Simon, R. The use of genomics in clinical trial design. Clin. Cancer Res.14, 5984–5993 (2008). - PubMed
MeSH terms
Substances
Grants and funding
- CEX2019-000891-S/Ministry of Economy and Competitiveness | Instituto de Salud Carlos III (Institute of Health Carlos III)
- ID 100010434; LCF/BQ/PR23/11980033/"la Caixa" Foundation (Caixa Foundation)
- LCF/BQ/DI22/11940038/"la Caixa" Foundation (Caixa Foundation)
- ID 100010434; LCF/BQ/DR21/11880009/"la Caixa" Foundation (Caixa Foundation)
- C14303/A17197, A19274/Cancer Research UK (CRUK)
LinkOut - more resources
Full Text Sources
Medical

