Multimodal deep learning for predicting neoadjuvant treatment outcomes in breast cancer: a systematic review
- PMID: 40551237
- PMCID: PMC12183913
- DOI: 10.1186/s13062-025-00661-8
Multimodal deep learning for predicting neoadjuvant treatment outcomes in breast cancer: a systematic review
Abstract
Background: Pathological complete response (pCR) to neoadjuvant systemic therapy (NAST) is an established prognostic marker in breast cancer (BC). Multimodal deep learning (DL), integrating diverse data sources (radiology, pathology, omics, clinical), holds promise for improving pCR prediction accuracy. This systematic review synthesizes evidence on multimodal DL for pCR prediction and compares its performance against unimodal DL.
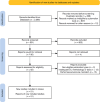
Methods: Following PRISMA, we searched PubMed, Embase, and Web of Science (January 2015-April 2025) for studies applying DL to predict pCR in BC patients receiving NAST, using data from radiology, digital pathology (DP), multi-omics, and/or clinical records, and reporting AUC. Data on study design, DL architectures, and performance (AUC) were extracted. A narrative synthesis was conducted due to heterogeneity.
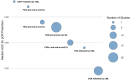
Results: Fifty-one studies, mostly retrospective (90.2%, median cohort 281), were included. Magnetic resonance imaging and DP were common primary modalities. Multimodal approaches were used in 52.9% of studies, often combining imaging with clinical data. Convolutional neural networks were the dominant architecture (88.2%). Longitudinal imaging improved prediction over baseline-only (median AUC 0.91 vs. 0.82). Overall, the median AUC across studies was 0.88, with 35.3% achieving AUC ≥ 0.90. Multimodal models showed a modest but consistent improvement over unimodal approaches (median AUC 0.88 vs. 0.83). Omics and clinical text were rarely primary DL inputs.
Conclusion: DL models demonstrate promising accuracy for pCR prediction, especially when integrating multiple modalities and longitudinal imaging. However, significant methodological heterogeneity, reliance on retrospective data, and limited external validation hinder clinical translation. Future research should prioritize prospective validation, integration underutilized data (multi-omics, clinical), and explainable AI to advance DL predictors to the clinical setting.
Keywords: Breast cancer; Deep learning; Multimodal prediction; Neoadjuvant treatment.
© 2025. The Author(s).
Conflict of interest statement
Declarations: Ethics approval and consent to participate. Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
- "Ricerca Corrente"/Ministero della Salute
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

