An ultrasensitive and specific CRISPR-Cas13a-mediated point-of-care assay for monkeypox detection and PCR-based clade detection
- PMID: 40551276
- PMCID: PMC12183853
- DOI: 10.1186/s40249-025-01325-5
An ultrasensitive and specific CRISPR-Cas13a-mediated point-of-care assay for monkeypox detection and PCR-based clade detection
Abstract
Background: The rapid increase in the number of monkeypox cases poses a considerable threat to the international community, necessitating sensitive, fast, and available diagnostic methods. Therefore, the objective of this study was to develop a rapid, sensitive and simple method with high clinical applicability.
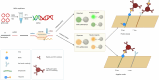
Methods: We developed a simple, rapid point-of-care assay to detect monkeypox virus (MPXV) using multienzyme isothermal rapid amplification (MIRA) coupled with the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas13a system. The detection system was optimized by synthesizing plasmids, and the detection sensitivity was explored by the continuous dilution of the plasmid. We validated the accuracy of this assay on 202 clinical MPXV samples and 104 interference samples through the kappa test. The visual interpretation of the results was realized by combining the assay with lateral flow strips. In addition, we developed a PCR-based method to identify MPXV Clades I and II, and the accuracy was tested through a kappa test on 202 clinical monkeypox samples and 104 interference samples.
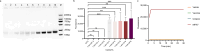
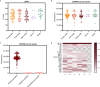
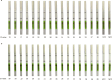
Results: Our assay achieved an analytical sensitivity of 14.4 copies/ml and high selectivity, as it differentiated MPXV from three other Orthopoxvirus species. The clinical testing results for 202 monkeypox samples and 104 interference samples demonstrated 100% sensitivity and specificity. Compared with quantitative PCR (qPCR), three samples tested as positive using our assay, which showed that the performance of this assay was superior to that of the qPCR assay. Combined with lateral flow strips, its availability and simplicity provide an alternative point-of-care diagnostic method for MPXV testing in remote settings and resource-poor areas. The results of 32 clinical samples showed that lateral flow strips had a high detection sensitivity and could identify samples with Ct value of 39 as positive. The clade identification assay detected as few as 200 copies/ml within 40 min and no cross-reaction was observed between Clades I and II. The clinical samples tested were all Clade II, which was consistent with the circulating clade in the Chinese mainland.
Conclusions: The MIRA-CRISPR-Cas13a-MPXV system offers a rapid, sensitive and specific approach for monkeypox diagnosis, with significance for monitoring monkeypox epidemics. The clade identification assay based on PCR could accurately distinguish Clade I from Clade II within 40 min and can be implemented for high-throughput operation.
Keywords: CRISPR-Cas13a system; Lateral flow strip; MPXV clade detection; Monkeypox; Point-of-care assay.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements. All samples used in this study were from the patient’s leftover swabs, plasma and serum after clinical inspection and did not involve the patient’s privacy. This study was approved by the Medical Ethics Committee of the Second Hospital of Nanjing (2024-LS-ky-113). The study was conducted in accordance with Declaration of Helsinki. Consent for publication: Not applicable. Competing interests: The authors AC and NZ were employed by the Suzhou TianLong Biotechnology Company Limited. The authors declare that they have no competing interests.
Figures

Similar articles
-
A Single-Tube Two-Step MIRA-CRISPR/Cas12b Assay for the Rapid Detection of Mpox Virus.Viruses. 2025 Jun 12;17(6):841. doi: 10.3390/v17060841. Viruses. 2025. PMID: 40573432 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Implementation of RT-RAA and CRISPR/Cas13a for an NiV Point-of-Care Test: A Promising Tool for Disease Control.Viruses. 2025 Mar 27;17(4):483. doi: 10.3390/v17040483. Viruses. 2025. PMID: 40284926 Free PMC article.
-
Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article.
-
[One-Step Detection of Human Influenza B Virus Through Recombinase Polymerase Amplification and CRISPR/Cas12a Protein].Sichuan Da Xue Xue Bao Yi Xue Ban. 2025 Mar 20;56(2):549-555. doi: 10.12182/20250360105. Sichuan Da Xue Xue Bao Yi Xue Ban. 2025. PMID: 40599263 Free PMC article. Chinese.
References
-
- Gessain A, Nakoune E, Yazdanpanah Y. Monkeypox. N Engl J Med. 2022;387(19):1783–93. - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

