Targeting SLFN11-regulated pathways restores chemotherapy sensitivity in AML
- PMID: 40552140
- PMCID: PMC12182837
- DOI: 10.1016/j.bneo.2024.100037
Targeting SLFN11-regulated pathways restores chemotherapy sensitivity in AML
Abstract
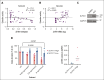
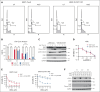
Chemoresistance represents an ongoing challenge in treating patients with acute myeloid leukemia (AML), and a better understanding of the resistance mechanisms can lead to the development of novel AML therapies. Here, we demonstrated that low expression of the DNA damage response gene Schlafen 11 (SLFN11) correlates with poor overall survival and worse prognosis in patients with AML. Moreover, we showed that SLFN11 plays an essential role in regulating chemotherapy sensitivity in AML. AML cells with suppressed levels of SLFN11 do not undergo apoptosis in response to cytarabine because of aberrant activation of the Ataxia telangiectasia and Rad3-related protein (ATR)/Checkpoint kinase 1 (Chk1) pathway, allowing for DNA damage repair, whereas sensitivity to cytarabine can be restored by inhibiting the ATR pathway. Importantly, SLFN11 knockout AML cells retain sensitivity to hypomethylating agents and the B-cell lymphoma 2 (BCL-2) inhibitor venetoclax. Altogether, these results reveal SLFN11 as an important regulator and predictor of chemotherapy sensitivity in AML and suggest that targeting pathways suppressed by SLFN11 may offer potential combination therapies to enhance and optimize chemotherapy responses in AML.
© 2024 by The American Society of Hematology. Licensed under Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International (CC BY-NC-ND 4.0), permitting only noncommercial, nonderivative use with attribution. All other rights reserved.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures





References
-
- Surveillance, Epidemiology, and End Results (SEER) Program . 2021. Acute myeloid leukemia - cancer stat facts: @NCICancerStats.https://seer.cancer.gov/statfacts/html/amyl.html
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

