Molecular Characterization of Plasmid-Mediated Quinolone Resistance Genes in Multidrug-Resistant Escherichia coli Isolated From Wastewater Generated From the Hospital Environment
- PMID: 40552214
- PMCID: PMC12183395
- DOI: 10.1177/11786302251342936
Molecular Characterization of Plasmid-Mediated Quinolone Resistance Genes in Multidrug-Resistant Escherichia coli Isolated From Wastewater Generated From the Hospital Environment
Abstract
Aim: This study investigated the carriage of Plasmid-Mediated Quinolone Resistance (PMQR) genes in fluoroquinolone-resistant Escherichia coli recovered from wastewater generated by healthcare institutions.
Materials and methods: Isolation of fluoroquinolone-resistant Escherichia coli was done on medium supplemented with 1 µg/mL of ciprofloxacin (a fluoroquinolone). Presumptive isolates were identified via the detection of uidA gene. Susceptibility of the isolates to a panel of antibiotics was done using disc diffusion method. Detection of PMQR genes in the isolates was done using primer-specific PCR.
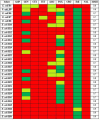
Results: Thirty fluoroquinolone-resistant Escherichia coli were obtained from the wastewater over a period of 6 months. The resistance to each of the antibiotic tested was: ampicillin (100%), ceftriaxone (100%), nalidixic acid (100%), tetracycline (96.7%), cefotaxime (96.7%), amoxicillin-clavulanate (80%), gentamicin (60%), cefoxitin (30%), and imipenem (3.3%). The Multiple Antibiotic Resistance Index (MARI) ranged from 0.6 to 0.9. The detection of PMQR genes in the 30 isolates was: qnrA (76.7%), qnrB (53.3%), qnrS (63.3%), aac(6')-lb-cr (43.3%), and qepA (43.3%). All the fluoroquinolone-resistant Escherichia coli carried at least one PMQR determinant.
Conclusion: This study revealed that untreated hospital wastewaters are significant hub of multidrug-resistant and fluoroquinolone-resistant Escherichia coli, showing high carriage of PMQR genes, and may be a major contributor to the resistome of fluoroquinolone-resistant bacteria in the Nigerian environment.
Keywords: Fluoroquinolone-resistant Escherichia coli; Multiple Antibiotic Resistance Index (MARI); Plasmid-Mediated Quinolone Resistance (PMQR); antibiotic resistance; healthcare settings.
Plain language summary
Escherichia coli showing resistance ciprofloxacin, a fluoroquinolone antibiotic was isolated from wastewater of some selected healthcare institutions in South-west Nigeria. The bacteria were identified using the detection of uidA gene, and they were tested against some antibiotics using the disc diffusion test. The detection of PMQR genes was done using primer-specific PCR. A total of 30 fluoroquinolone-resistant isolates were recovered from the wastewater in six months. All the isolates were resistant to ampicillin and nalidixic acid, while the least level of resistance was to imipenem, a carbapenem antibiotic. The multiple antibiotic resistance index for the isolates ranged from 0.6 to 0.9. All the isolates were multidrug-resistant. The frequency of PMQR genes in the 30 resistant isolates obtained from the wastewater was: qnrA (76.7%), qnrS (60%), qnrB (53%), aac(6')-lb-cr (43.3%) and qepA (40%). All the isolates carried at least one of the target PMQR genes. Wastewater from healthcare institution is a breeding ground for fluoroquinolone-resistant Escherichia coli.
© The Author(s) 2025.
Conflict of interest statement
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures


References
-
- Aminul P, Anwar S, Molla MMA, Miah MRA. Evaluation of antibiotic resistance patterns in clinical isolates of Klebsiella pneumoniae in Bangladesh. Biosaf Heal. 2021;3:301-306.
LinkOut - more resources
Full Text Sources

