Evolutionary insights into toxins diversity in Ceriantharia (Cnidaria; Anthozoa)
- PMID: 40552252
- PMCID: PMC12182779
- DOI: 10.1016/j.toxcx.2025.100227
Evolutionary insights into toxins diversity in Ceriantharia (Cnidaria; Anthozoa)
Abstract
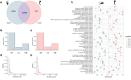
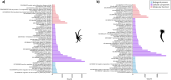
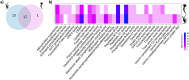
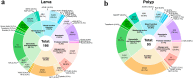
Ceriantharians synthesize and inoculate the toxins found in their stinging cells spread throughout the body. For most cnidarians the putative toxins profile can vary widely depending on the tissue function and the environmental conditions faced by these marine invertebrates. Extensive gene duplications events have impacted the diversity of the toxins system of cnidarians and could explain the rapid emergence of novel toxins. On the other hand, it seems for Ceriantharia, the putative toxins profile does not exhibit major variation, despite occupying different ecological niches. Some species of ceriantharians have a planktonic stage that is highly dispersive, while the benthic phase is characterized by semi-sessile polyp. However, the polyp builds a tube involving the entire column that can play an additional function by protecting against predators and competitors, which could decrease the need to synthesize a wide array of toxins. In the present study, we compare the putative toxins of the larva and polyp of Arachnanthus errans based on the functional annotations of the transcriptomes against annotated protein databases. We seek to understand the evolutionary process of two toxin-like protein families using phylogenetic reconstruction methods with target sequences of the transcriptome of nine ceriantharian species. Our exploration revealed that the larva expresses 70 more toxin-like genes than the polyp, which may relate to abiotic and biotic factors the larva experiences. Our phylogenetic analyses suggest duplication events may have occurred in both toxins-like proteins and the two copies of Kunitz-like proteins might have been present in the common ancestor of Ceriantharia.
Keywords: Development stages; Evolution; Functional genomics; RNASeq; Toxin-like genes; Tube-dwelling anemones.
© 2025 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Andrews S. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics. 2010 http://www.bioinformatics.babraham.ac.uk/projects/fastqc/GoogleScholar Retrieved from.
-
- Arai M.N. Predation on pelagic coelenterates: a review. J. Mar. Biol. Assoc. U. K. 2005;85:523–536. doi: 10.1017/S0025315405011458. - DOI
LinkOut - more resources
Full Text Sources

