ELOVL2 mediated stabilization of AR contributes to enzalutamide resistance in prostate cancer
- PMID: 40552308
- PMCID: PMC12183063
- DOI: 10.3389/fcell.2025.1598400
ELOVL2 mediated stabilization of AR contributes to enzalutamide resistance in prostate cancer
Erratum in
-
Correction: ELOVL2 mediated stabilization of AR contributes to enzalutamide resistance in prostate cancer.Front Cell Dev Biol. 2025 Jun 25;13:1646699. doi: 10.3389/fcell.2025.1646699. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40636670 Free PMC article.
Abstract
Introduction: To investigate the molecular mechanisms underlying enzalutamide resistance in castration-resistant prostate cancer (CRPC) and explore potential therapeutic strategies to overcome resistance.
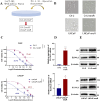
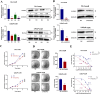
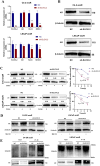
Methods: We conducted comprehensive bioinformatic analysis using LNCaP/enzalutamide-resistant cells to identify key pathways associated with resistance. Functional validation was performed through targeted inhibition of the elongation of very-long chain fatty acid protein 2 (ELOVL2), followed by assays to assess cancer cell proliferation and enzalutamide sensitivity. Mechanistic studies were conducted to evaluate the impact of ELOVL2 on the ubiquitin-proteasome system and AR signaling pathways.
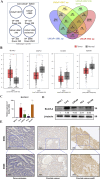
Results: Bioinformatic analysis revealed that activation of fatty acid metabolism, particularly through upregulation of ELOVL2, plays a critical role in driving enzalutamide resistance in PCa. Functional studies demonstrated that targeted inhibition of ELOVL2 significantly suppressed cancer cell proliferation and restored enzalutamide sensitivity in resistant cells. Mechanistically, ELOVL2 facilitates enzalutamide resistance by impairing the ubiquitin-proteasome system, leading to the subsequent activation of AR signaling pathways.
Discussion: Our findings demonstrate that ELOVL2 drives enzalutamide resistance in CRPC by stabilizing AR through inhibition of ubiquitin-proteasome-mediated degradation. Targeting ELOVL2 represents a promising therapeutic strategy to overcome resistance in CRPC, with potential to improve clinical outcomes for patients.
Keywords: CRPC; ELOVL2; androgen receptor; enzalutamide resistance; prostate cancer.
Copyright © 2025 Cen, Guo, Zeng, Song, Ge, Chen, Li, Yu, Lv and Zhao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Reciprocal regulation between RACGAP1 and AR contributes to endocrine therapy resistance in prostate cancer.Cell Commun Signal. 2024 Jun 19;22(1):339. doi: 10.1186/s12964-024-01703-w. Cell Commun Signal. 2024. PMID: 38898473 Free PMC article.
-
Cost-effectiveness of enzalutamide with androgen-deprivation therapy (ADT) versus ADT alone for the treatment of high-risk biochemically recurrent non-metastatic castration-sensitive prostate cancer in Canada.J Med Econ. 2025 Dec;28(1):766-777. doi: 10.1080/13696998.2025.2503660. Epub 2025 May 23. J Med Econ. 2025. PMID: 40395149
-
Comparative efficacy of second-generation androgen receptor inhibitors for treating prostate cancer: A systematic review and network meta-analysis.Front Endocrinol (Lausanne). 2023 Mar 9;14:1134719. doi: 10.3389/fendo.2023.1134719. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36967752 Free PMC article.
-
The efficacy and adverse events of conventional and second-generation androgen receptor inhibitors for castration-resistant prostate cancer: A network meta-analysis.Front Endocrinol (Lausanne). 2023 Feb 10;14:1131033. doi: 10.3389/fendo.2023.1131033. eCollection 2023. Front Endocrinol (Lausanne). 2023. PMID: 36843606 Free PMC article.
-
Castration-resistant prostate cancer is resensitized to androgen deprivation by autophagy-dependent apoptosis induced by blocking SKP2.Sci Signal. 2024 Dec 17;17(867):eadk4122. doi: 10.1126/scisignal.adk4122. Epub 2024 Dec 17. Sci Signal. 2024. PMID: 39689183 Free PMC article.
References
-
- Chan S. C., Selth L. A., Li Y., Nyquist M. D., Miao L., Bradner J. E., et al. (2015). Targeting chromatin binding regulation of constitutively active AR variants to overcome prostate cancer resistance to endocrine-based therapies. Nucleic Acids Res. 43 (12), 5880–5897. 10.1093/nar/gkv262 - DOI - PMC - PubMed
-
- Choudhury D., Dolezal J. M., Dyer E., Kochanny S., Ramesh S., Howard F. M., et al. (2024). Developing a low-cost, open-source, locally manufactured workstation and computational pipeline for automated histopathology evaluation using deep learning. eBioMedicine 107, 105276. 10.1016/j.ebiom.2024.105276 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

