Phenotypic analysis of various Clostridioides difficile ribotypes reveals consistency among core processes
- PMID: 40552806
- PMCID: PMC12285255
- DOI: 10.1128/aem.00964-25
Phenotypic analysis of various Clostridioides difficile ribotypes reveals consistency among core processes
Abstract
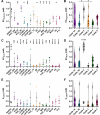
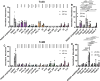
Clostridioides difficile infections (CDI) cause almost 300,000 hospitalizations per year, of which ~15%-30% are the result of recurring infections. The prevalence and persistence of CDI in hospital settings have resulted in an extensive collection of C. difficile clinical isolates and their classification, typically by ribotype. While much of the current literature focuses on one or two prominent epidemic ribotypes (e.g., RT027), recent years have seen several other ribotypes dominate the clinical landscape (e.g., RT106 and RT078). Some ribotypes are associated with severe disease and/or increased recurrence rates, but why certain ribotypes are more prominent or harmful than others remains unknown. Because C. difficile has a large, open pan-genome, this observed relationship between ribotype and clinical outcome could be a result of the genetic diversity of C. difficile. Thus, we hypothesize that the core biological processes of C. difficile are conserved across ribotypes/clades. We tested this hypothesis by observing the growth kinetics, sporulation, germination, production of toxin A and toxin B, bile acid sensitivity, bile salt hydrolase activity, and surface motility of 15 strains belonging to various ribotypes spanning each known C. difficile clade. In viewing these phenotypes across each strain, we see that core phenotypes (growth, germination, sporulation, and resistance to bile salt toxicity) are remarkably consistent across clades/ribotypes. This suggests that variations observed in the clinical setting may be due to unidentified factors in the accessory genome or due to unknown host factors.IMPORTANCEClostridioides difficile infections impact thousands of individuals every year, many of whom experience recurring infections. Clinical studies have reported an unexplained correlation between some clades/ribotypes of C. difficile and disease severity/recurrence. Here, we demonstrate that C. difficile strains across major clades/ribotypes are consistent in their core phenotypes. This suggests that such phenotypes are not responsible for variations in disease severity/recurrence and are ideal targets for the development of therapeutics meant to treat C. difficile-related infections.
Keywords: Clostridium difficile; clade; phenotypes; physiology; ribotype.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
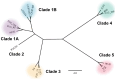
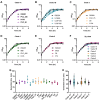
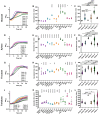
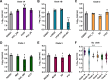


Update of
-
Phenotypic analysis of various Clostridioides difficile ribotypes reveals consistency among core processes.bioRxiv [Preprint]. 2025 Jan 10:2025.01.10.632434. doi: 10.1101/2025.01.10.632434. bioRxiv. 2025. Update in: Appl Environ Microbiol. 2025 Jul 23;91(7):e0096425. doi: 10.1128/aem.00964-25. PMID: 39829883 Free PMC article. Updated. Preprint.
Similar articles
-
Phenotypic analysis of various Clostridioides difficile ribotypes reveals consistency among core processes.bioRxiv [Preprint]. 2025 Jan 10:2025.01.10.632434. doi: 10.1101/2025.01.10.632434. bioRxiv. 2025. Update in: Appl Environ Microbiol. 2025 Jul 23;91(7):e0096425. doi: 10.1128/aem.00964-25. PMID: 39829883 Free PMC article. Updated. Preprint.
-
Characterization of Flagellum and Toxin Phase Variation in Clostridioides difficile Ribotype 012 Isolates.J Bacteriol. 2018 Jun 25;200(14):e00056-18. doi: 10.1128/JB.00056-18. Print 2018 Jul 15. J Bacteriol. 2018. PMID: 29735765 Free PMC article.
-
Whole genome sequence analysis reveals limited diversity among Clostridioides difficile ribotype 027 and 078 isolates collected in 22 hospitals in Berlin and Brandenburg, Germany.Antimicrob Resist Infect Control. 2025 May 28;14(1):56. doi: 10.1186/s13756-025-01565-y. Antimicrob Resist Infect Control. 2025. PMID: 40437588 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Fecal microbiota transplantation for the treatment of recurrent Clostridioides difficile (Clostridium difficile).Cochrane Database Syst Rev. 2023 Apr 25;4(4):CD013871. doi: 10.1002/14651858.CD013871.pub2. Cochrane Database Syst Rev. 2023. PMID: 37096495 Free PMC article.
Cited by
-
Unique growth and morphology properties of Clade 5 Clostridioides difficile strains revealed by single-cell time-lapse microscopy.bioRxiv [Preprint]. 2025 May 16:2024.02.13.580212. doi: 10.1101/2024.02.13.580212. bioRxiv. 2025. Update in: PLoS Pathog. 2025 May 21;21(5):e1013155. doi: 10.1371/journal.ppat.1013155. PMID: 40463200 Free PMC article. Updated. Preprint.
References
-
- Kochan TJ, Shoshiev MS, Hastie JL, Somers MJ, Plotnick YM, Gutierrez-Munoz DF, Foss ED, Schubert AM, Smith AD, Zimmerman SK, Carlson PE Jr, Hanna PC. 2018. Germinant synergy facilitates Clostridium difficile spore germination under physiological conditions. mSphere 3:e00335-18. doi: 10.1128/mSphere.00335-18 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

