Evidence that hPIV2 paramyxovirus antigenomes are edited during infection
- PMID: 40552829
- PMCID: PMC12345181
- DOI: 10.1128/mbio.03667-24
Evidence that hPIV2 paramyxovirus antigenomes are edited during infection
Abstract
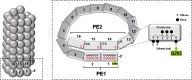
Mononegavirus genomes contain cis-acting sequences that direct the formation of mRNA 5' and 3' ends during synthesis (gene start [GS] and gene end [GE]) but are silent during antigenome synthesis from the same genome template. Paramyxo- and filoviruses also carry an additional cis-acting sequence (EDIT) that directs the insertion of specific nucleotide during mRNA synthesis, expanding gene products. This "mRNA editing" is associated with viruses whose replication is governed by the rule of six. One might, therefore, assume that EDIT, like GS and GE, would be restricted to that of mRNA synthesis. However, accurate determination of the ratio of edited to unedited RNAs in different sub-populations of wild-type and mutant human parainfluenza virus type 2 (hPIV2) infections finds evidence that antigenomes are also edited during infection. This alters our view of how the rule of six governs mononegavirus infections and why this rule, RNA editing, and bipartite promoters are linked.
Importance: We have assumed that paramyxovirus editing signals would operate strictly during mRNA synthesis, as it apparently makes no sense to edit antigenomes. Nevertheless, there is evidence here that the opposite is the case. If so, this alters our view of paramyxovirus replication, and we summarize what is known about how its RNA-dependent RNA polymerase carries out its task of expressing alternate open reading frames during mRNA synthesis.
Keywords: RNA editing; paramyxovirus; rule of six; viral replication.
Conflict of interest statement
Yusuke Matsumoto receives compensation from Denka Co., Ltd. The other authors declare no competing interest.
Figures
References
-
- Desfosses A, Milles S, Jensen MR, Guseva S, Colletier JP, Maurin D, Schoehn G, Gutsche I, Ruigrok RWH, Blackledge M. 2019. Assembly and cryo-EM structures of RNA-specific measles virus nucleocapsids provide mechanistic insight into paramyxoviral replication. Proc Natl Acad Sci USA 116:4256–4264. doi: 10.1073/pnas.1816417116 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

