PVGA: a precise viral genome assembler using an iterative alignment graph
- PMID: 40552980
- PMCID: PMC12206156
- DOI: 10.1093/gigascience/giaf063
PVGA: a precise viral genome assembler using an iterative alignment graph
Abstract
Background: Viral genome analysis is crucial for understanding virus evolution and mutation. Investigations into viral evolutionary dynamics and mutation patterns have garnered significant research attention since the outbreak of COVID-19. The basic structure of many virus genomes is highly conserved [1]. RNA viruses have high mutation rates, and single-nucleotide variations may induce substantial phenotypic alterations in terms of viral function and pathogenicity. Thus, special assembly methods are required for viral genome analysis.
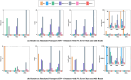
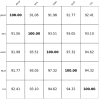
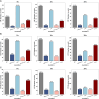
Result: PVGA starts with a reference genome and the sequencing reads. The first step in PVGA involves constructing an alignment graph based on a reference genome and the set of input sequencing reads. Then the optimal genomic path is determined through dynamic programming, maximizing the cumulative edge weights that reflect read support density across the alignment graph. The obtained path corresponds to a refined genome. Finally, we repeat the process by using the new reference genomes until no further improvement is possible. We evaluate PVGA's performance across both assembly and polishing tasks using simulated and real datasets, including both long reads and short reads. The experiments demonstrate that PVGA always outperforms popular existing programs in terms of the quality of assembly results, while the running time of our method is compatible to others. In particular, simulated Nanopore datasets show that our method can correctly report the true genomes with 0 mismatches and 0 indels.
Conclusions: PVGA is a novel viral genome assembler that seamlessly integrates assembly and polishing into a unified workflow. Its design prioritizes high accuracy, enabling the detection of subtle genomic variations that can impact viral function and pathogenicity. By addressing the unique challenges of viral genome assembly, PVGA provides a reliable and precise solution for advancing our understanding of viral evolution and behavior.
Keywords: alignment graph; genome assembler; iterative method; maximum total weight path; virus genome.
© The Author(s) 2025. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Lessons learned: overcoming common challenges in reconstructing the SARS-CoV-2 genome from short-read sequencing data via CoVpipe2.F1000Res. 2024 Apr 16;12:1091. doi: 10.12688/f1000research.136683.2. eCollection 2023. F1000Res. 2024. PMID: 38716230 Free PMC article.
-
Enhancing public health surveillance: a comparative study of platform-specific and hybrid assembly approaches in SARS-CoV-2 genome sequencing.Microb Genom. 2025 Jul;11(7):001357. doi: 10.1099/mgen.0.001357. Microb Genom. 2025. PMID: 40637372 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Application of a high-resolution melt assay for monitoring SARS-CoV-2 variants in Burkina Faso and Kenya.mSphere. 2025 Jun 25;10(6):e0002725. doi: 10.1128/msphere.00027-25. Epub 2025 May 29. mSphere. 2025. PMID: 40439429 Free PMC article.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

