Lung adenocarcinoma-derived IFN-γ promotes growth by modulating CD8+ T cell production of CCR5 chemokines
- PMID: 40553564
- PMCID: PMC12404755
- DOI: 10.1172/JCI191070
Lung adenocarcinoma-derived IFN-γ promotes growth by modulating CD8+ T cell production of CCR5 chemokines
Abstract
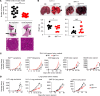
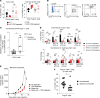
Because the lung is a mucosal barrier organ with a unique immunologic environment, mechanisms of immunoregulation in lung cancer may differ from those of other malignancies. Consistent with this notion, we found that CD8+ T cells played a paradoxical role in facilitating, rather than ameliorating, the growth of multiple lung adenocarcinoma models. These included spontaneous, carcinogen-induced, and transplantable tumor cell line models. Specifically, we found that CD8+ T cells promoted homing of CD4+Foxp3+ Tregs to the tumor bed by increasing the levels of CCR5 chemokines in the tumor microenvironment in an IFN-γ- and TNF-α-dependent manner. Contrary to their canonical role, these Th1 cytokines contributed to accelerated growth of murine lung adenocarcinomas, while suppressing the growth of other malignancies. Surprisingly, lung cancer cells themselves can serve as a dominant source of IFN-γ, and deletion of this cytokine from cancer cells using CRISPR/Cas9 decreases tumor growth. Importantly for translational applications, in patients with lung cancer, a high level of IFN-γ was also found at both the mRNA and protein levels. Our data outline what we deem a novel and previously undefined lung cancer-specific immunoregulatory pathway that may be harnessed to tailor immune-based therapy specifically for this malignancy.
Keywords: Adaptive immunity; Cell biology; Immunology; Lung cancer; Oncology; T cells.
Conflict of interest statement
Figures




References
-
- Yee C, et al. Adoptive T cell therapy using antigen-specific CD8+ T cell clones for the treatment of patients with metastatic melanoma: in vivo persistence, migration, and antitumor effect of transferred T cells. Proc Natl Acad Sci U S A. 2002;99(25):16168–16173. doi: 10.1073/pnas.242600099. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

