BET inhibitors reduce tumor growth in preclinical models of gastrointestinal gene signature-positive castration-resistant prostate cancer
- PMID: 40553565
- PMCID: PMC12352905
- DOI: 10.1172/JCI180378
BET inhibitors reduce tumor growth in preclinical models of gastrointestinal gene signature-positive castration-resistant prostate cancer
Abstract
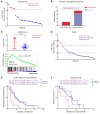
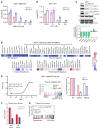
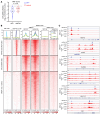
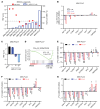
A subgroup (~20%-30%) of castration-resistant prostate cancer (CRPC) aberrantly expresses a gastrointestinal (GI) transcriptome governed by 2 GI-lineage-restricted transcription factors, HNF1A and HNF4G. In this study, we found that expression of GI transcriptome in CRPC correlated with adverse clinical outcomes to androgen receptor (AR) signaling inhibitor treatment and shorter overall survival. Bromo- and extraterminal domain inhibitors (BETi) downregulated HNF1A, HNF4G, and the GI transcriptome in multiple CRPC models, including cell lines, patient-derived organoids, and patient-derived xenografts, whereas AR and the androgen-dependent transcriptome were largely spared. Accordingly, BETi selectively inhibited growth of GI transcriptome-positive preclinical models of prostate cancer. Mechanistically, BETi inhibited BRD4 binding at enhancers globally, including both AR and HNF4G bound enhancers, while gene expression was selectively perturbed. Restoration of HNF4G expression in the presence of BETi rescued target gene expression without rescuing BRD4 binding. This suggests that inhibition of master transcription factors expression underlies the selective transcriptional effects of BETi.
Keywords: Cell biology; Drug therapy; Epigenetics; Genetics; Oncology.
Conflict of interest statement
Figures


Update of
-
BET inhibitors as a therapeutic intervention in gastrointestinal gene signature-positive castration-resistant prostate cancer.bioRxiv [Preprint]. 2024 Mar 13:2024.03.09.584256. doi: 10.1101/2024.03.09.584256. bioRxiv. 2024. Update in: J Clin Invest. 2025 Jun 24;135(16):e180378. doi: 10.1172/JCI180378. PMID: 38559135 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

