Nemo-like kinase disrupts nuclear import and drives TDP43 mislocalization in ALS
- PMID: 40553568
- PMCID: PMC12404744
- DOI: 10.1172/JCI188138
Nemo-like kinase disrupts nuclear import and drives TDP43 mislocalization in ALS
Abstract
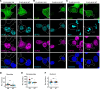
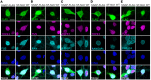
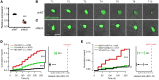
Cytoplasmic transactive response DNA-binding protein 43 (TDP43) mislocalization and aggregation are pathological hallmarks of amyotrophic lateral sclerosis (ALS). However, the initial cellular insults that lead to TDP43 mislocalization remain unclear. In this study, we demonstrate that nemo-like kinase (NLK) - a proline-directed serine-threonine kinase - promotes the mislocalization of TDP43 and other RNA-binding proteins by disrupting nuclear import. NLK levels were selectively elevated in neurons exhibiting TDP43 mislocalization in tissues from patients with ALS, and genetic reduction of NLK reduced toxicity in human neuron models of ALS. Our findings suggest that NLK is a promising therapeutic target for neurodegenerative diseases.
Keywords: ALS; Cell biology; Molecular pathology; Neuroscience; Transport.
Figures





Update of
-
Nemo-like kinase disrupts nuclear import and drives TDP43 mislocalization in ALS.bioRxiv [Preprint]. 2025 Jan 28:2025.01.27.635090. doi: 10.1101/2025.01.27.635090. bioRxiv. 2025. Update in: J Clin Invest. 2025 Jun 24;135(17):e188138. doi: 10.1172/JCI188138. PMID: 39975323 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

