MUC1-C dependence in treatment-resistant prostate cancer uncovers a target for antibody-drug conjugate therapy
- PMID: 40553569
- PMCID: PMC12288968
- DOI: 10.1172/jci.insight.190924
MUC1-C dependence in treatment-resistant prostate cancer uncovers a target for antibody-drug conjugate therapy
Abstract
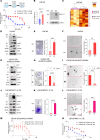
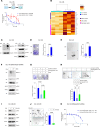
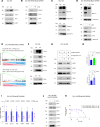
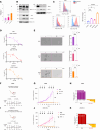
Androgen receptor-positive prostate cancer (PC), castration-resistant prostate cancer (CRPC), and neuroendocrine prostate cancer (NEPC) invariably become resistant to treatment with targeted and cytotoxic agents. Multiple pathways have been identified as being responsible for these pleiotropic mechanisms of resistance. The mucin 1 (MUC1) gene is aberrantly expressed in CRPC/NEPC in association with poor clinical outcomes; however, it is not known if the oncogenic MUC1-C/M1C protein drives treatment resistance. We demonstrated that MUC1-C is necessary for resistance of (i) PC cells to enzalutamide (ENZ) and (ii) CRPC and NEPC cells to docetaxel (DTX). Our results showed that MUC1-C-mediated resistance is conferred by upregulation of aerobic glycolysis and suppression of reactive oxygen species necessary for self-renewal. Dependence of these resistant phenotypes on MUC1-C for the cancer stem cell (CSC) state identified a potential target for treatment. In this regard, we further demonstrated that targeting MUC1-C with an M1C antibody-drug conjugate (ADC) is highly effective in suppressing (i) self-renewal of drug-resistant CRPC/NEPC CSCs and (ii) growth of treatment-emergent NEPC tumor xenografts derived from drug-resistant cells and a patient with refractory disease. These findings uncovered a common MUC1-C-dependent pathway in treatment-resistant CRPC/NEPC progression and identified MUC1-C as a target for their therapy with an M1C ADC.
Keywords: Cancer; Drug therapy; Oncology; Therapeutics; Urology.
Conflict of interest statement
Figures




References
-
- Seton-Rogers S. Prostate cancer: connecting androgen receptor and immunity. Nat Rev Cancer. 2016;16(5):273. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

