Identification of miR-106b-5p as a senolytic miRNA
- PMID: 40554391
- PMCID: PMC12219373
- DOI: 10.1016/j.ebiom.2025.105810
Identification of miR-106b-5p as a senolytic miRNA
Abstract
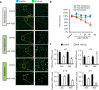
Background: Cellular senescence contributes to ageing and age-related diseases. While miR-106b-5p is elevated in centenarians and GH-deficient models of healthy ageing, its role in senescence was unclear.
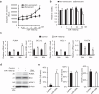
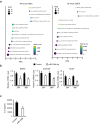
Methods: Senolytic effects of miR-106b-5p were evaluated in etoposide-induced senescent IMR90 fibroblasts and HUVECs, and in male naturally aged mice using liposome-mediated delivery. Cellular assays, qPCR, Western blotting, and RNA-seq were performed to assess senescence and SASP markers, apoptosis pathways, and molecular mechanisms.
Findings: miR-106b-5p selectively eliminated senescent cells without affecting non-senescent cells. It enhanced p53 K120 acetylation and upregulated PUMA, while reducing PCAF expression. In male aged mice, systemic delivery of miR-106b-5p reduced markers of senescence and SASP in multiple tissues and lowered serum IL-6 levels.
Interpretation: miR-106b-5p functions as a senolytic miRNA via modulation of the p53-PUMA axis and SASP suppression. It holds promise as a therapeutic agent to mitigate age-related cellular dysfunction and inflammation.
Funding: Supported by NIH (U19 AG056278, R01 AG063543, P01 AG062413, U54 AG079754, U54 AG076041, R01 AG069819, P01 AI172501), the Glenn Foundation, and NSF grant #2317758.
Keywords: Ageing and inflammation; Cellular senescence; SASP; Senolytic therapy; miR-106b-5p; p53-PUMA axis.
Copyright © 2025 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests Tianpeng Zhang, Allancer Nunes, Ryan O'Kelly, Luise Angelini, Michal M. Masternak, and Yousin Suh declare no competing interests related to this study. Laura J. Niedernhofer holds equity in Unity Biotechnology and has pending IP through the University of Minnesota related to senescence. Paul D. Robbins holds equity in Genascence Inc. and Innate Biologics, stock options in L&J Bio, and has pending patents on senotherapeutics via the University of Minnesota. Xiao Dong is a co-founder and shareholder of SingulOmics Corp. None of these disclosures are related to the present work, and no authors intend to file a patent based on this study.
Figures



References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

