Single-cell and spatiotemporal profile of ovulation in the mouse ovary
- PMID: 40554460
- PMCID: PMC12186953
- DOI: 10.1371/journal.pbio.3003193
Single-cell and spatiotemporal profile of ovulation in the mouse ovary
Abstract
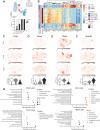
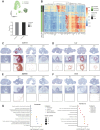
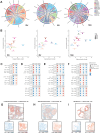
Ovulation is a spatiotemporally coordinated process that involves several tightly controlled events, including oocyte meiotic maturation, cumulus expansion, follicle wall rupture and repair, and ovarian stroma remodeling. To date, no studies have detailed the precise window of ovulation at single-cell resolution. Here, we performed parallel single-cell RNA-seq and spatial transcriptomics on paired mouse ovaries across an ovulation time course to map the spatiotemporal profile of ovarian cell types. We show that major ovarian cell types exhibit time-dependent transcriptional states enriched for distinct functions and have specific localization profiles within the ovary. We also identified gene markers for ovulation-dependent cell states and validated these using orthogonal methods. Finally, we performed cell-cell interaction analyses to identify ligand-receptor pairs that may drive ovulation, revealing previously unappreciated interactions. Taken together, our data provides a rich and comprehensive resource of murine ovulation that can be mined for discovery by the scientific community.
Copyright: © 2025 Huang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: A.K.S reports compensation for consulting and/or SAB membership from Honeycomb Biotechnologies, Cellarity, Ochre Bio, Relation Therapeutics, Bio-Rad Laboratories, Passkey Therapeutics, Fog Pharma, Dahlia Biosciences, and intrECate Biotherapeutics. B.A.G., R.H., C.E.K., Y.Z., S.L.F., C.M., A.K.S., F.E.D., H.C.L., L.T.Z., E.Z., and J.N. have filed a patent related to the work described in this study (patent application no. PCT/US24/25824).
Figures



Update of
-
Single-cell and spatiotemporal profile of ovulation in the mouse ovary.bioRxiv [Preprint]. 2024 Jun 5:2024.05.20.594719. doi: 10.1101/2024.05.20.594719. bioRxiv. 2024. Update in: PLoS Biol. 2025 Jun 24;23(6):e3003193. doi: 10.1371/journal.pbio.3003193. PMID: 38826447 Free PMC article. Updated. Preprint.
References
-
- Qublan H, Amarin Z, Nawasreh M, Diab F, Malkawi S, Al-Ahmad N, et al. Luteinized unruptured follicle syndrome: incidence and recurrence rate in infertile women with unexplained infertility undergoing intrauterine insemination. Hum Reprod. 2006;21(8):2110–3. doi: 10.1093/humrep/del113 . - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

