Evaluation of subretinally delivered Cas9 ribonucleoproteins in murine and porcine animal models highlights key considerations for therapeutic translation of genetic medicines
- PMID: 40554553
- PMCID: PMC12186880
- DOI: 10.1371/journal.pone.0317387
Evaluation of subretinally delivered Cas9 ribonucleoproteins in murine and porcine animal models highlights key considerations for therapeutic translation of genetic medicines
Abstract
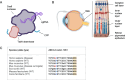
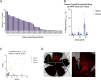
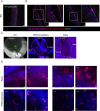
Genetic medicines, including CRISPR/Cas technologies, extend tremendous promise for addressing unmet medical need in inherited retinal disorders and other indications; however, there remain challenges for the development of therapeutics. Herein, we evaluate genome editing by engineered Cas9 ribonucleoproteins (eRNP) in vivo via subretinal administration using mouse and pig animal models. Subretinal administration of adenine base editor and double strand break-inducing Cas9 nuclease eRNPs mediate genome editing in both species. Editing occurs in retinal pigmented epithelium (RPE) and photoreceptor cells, with favorable tolerability in both species. Using transgenic reporter strains, we determine that editing primarily occurs close to the site of administration, within the bleb region associated with subretinal injection. Our results show that subretinal administration of BE-eRNPs in mice mediates base editing of up to 12% of the total neural retina, with an average rate of 7% observed at the highest dose tested. In contrast, a substantially lower editing efficiency was observed in minipigs; even with direct quantification of only the treated region, a maximum base editing rate of 1.5%, with an average rate of <1%, was observed. Our data highlight the importance of species consideration in preclinical studies for the development of genetic medicines targeting the eye and provide an example of a lack of translation between small and larger animal models in the context of subretinal administration of Cas9 eRNPs.
Copyright: © 2025 Wei et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
All authors except PKS, BRP, DG, and KS are employees of Spotlight Therapeutics, Inc., or were employees at the time the research took place. RM, KM, SCW, BGG, and MJJ are inventors on patent applications related to delivery of Cas9 ribonucleoproteins (PCT/US2020/024249 and PCT/US2023/076149). SCW is a prior consultant for BioEntre, consultant for Actym Therapeutics, an inventor on a patent for a mouse model of autoimmune adverse events (PCT/US2019/050551), and employee of Inversion Therapeutics. Benjamin Gowen is an employee of Editpep, Inc. KS is a member of the scientific advisory boards of Andson Biotech, Bharat Biotech, and Notch Therapeutics and is a board member for Andson Biotech and Bharat Biotech. This does not alter our adherence to the PLOS ONE policies on sharing data and materials.
Figures


Update of
-
Evaluation of subretinally delivered Cas9 ribonucleoproteins in murine and porcine animal models highlights key considerations for therapeutic translation of genetic medicines.bioRxiv [Preprint]. 2024 Dec 30:2024.12.30.630799. doi: 10.1101/2024.12.30.630799. bioRxiv. 2024. Update in: PLoS One. 2025 Jun 24;20(6):e0317387. doi: 10.1371/journal.pone.0317387. PMID: 39803585 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

