Characterization of human CMV-specific CD8+ T cells using multi-layer single-cell omics
- PMID: 40555232
- PMCID: PMC12296424
- DOI: 10.1016/j.crmeth.2025.101085
Characterization of human CMV-specific CD8+ T cells using multi-layer single-cell omics
Abstract
In this study, we established a comprehensive workflow to collect multi-omics single-cell data using a commercially available micro-well-based platform. This included whole transcriptome, cell surface markers (targeted sequencing-based cell surface proteomics), T cell specificities, adaptive immune receptor repertoire (AIRR) profiles, and sample multiplexing. With this technique, we identified paired T cell receptor sequences for three prominent human CMV epitopes. In addition, we assessed the ability of dCODE dextramers to detect antigen-specific T cells at low frequencies by estimating sensitivities and specificities when used as reagents for single-cell multi-omics.
Keywords: BD Rhapsody; CD8(+) T cells; CMV; CP: immunology; TCR sequencing; antigen-specific T cells; dCODE dextramers; single-cell multi-omics; systems immunology.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


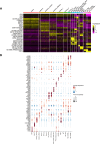
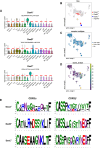

References
-
- Valkiers S., de Vrij N., Gielis S., Verbandt S., Ogunjimi B., Laukens K., Meysman P. Recent advances in T-cell receptor repertoire analysis: bridging the gap with multimodal single-cell RNA sequencing. ImmunoInformatics. 2022;5 doi: 10.1016/j.immuno.2022.100009. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

