In vivo self-renewal and expansion of quiescent stem cells from a non-human primate
- PMID: 40555729
- PMCID: PMC12187938
- DOI: 10.1038/s41467-025-58897-x
In vivo self-renewal and expansion of quiescent stem cells from a non-human primate
Abstract
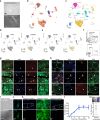
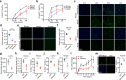
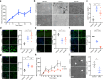
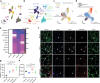
The development of non-human primate models is essential for the fields of developmental and regenerative biology because those models will more closely approximate human biology than do murine models. Based on single cell RNAseq and fluorescence-activated cell sorting, we report the identification and functional characterization of two quiescent stem cell populations (skeletal muscle stem cells (MuSCs) and mesenchymal stem cells termed fibro-adipogenic progenitors (FAPs)) in the non-human primate Microcebus murinus (the gray mouse lemur). We demonstrate in vivo proliferation, differentiation, and self-renewal of both MuSCs and FAPs. By combining cell phenotyping with cross-species molecular profiling and pharmacological interventions, we show that mouse lemur MuSCs and FAPs are more similar to human than to mouse counterparts. We identify unexpected gene targets involved in regulating primate MuSC proliferation and primate FAP adipogenic differentiation. Moreover, we find that the cellular composition of mouse lemur muscle better models human muscle than does macaque (Macaca fascicularis) muscle. Finally, we note that our approach presents as a generalizable pipeline for the identification, isolation, and characterization of stem cell populations in new animal models.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures
Update of
-
In vivo self-renewal and expansion of quiescent stem cells from a non-human primate.bioRxiv [Preprint]. 2025 Mar 28:2025.03.27.645793. doi: 10.1101/2025.03.27.645793. bioRxiv. 2025. Update in: Nat Commun. 2025 Jun 24;16(1):5370. doi: 10.1038/s41467-025-58897-x. PMID: 40196588 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

