Bifidobacterium deficit in United States infants drives prevalent gut dysbiosis
- PMID: 40555747
- PMCID: PMC12187928
- DOI: 10.1038/s42003-025-08274-7
Bifidobacterium deficit in United States infants drives prevalent gut dysbiosis
Abstract
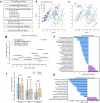
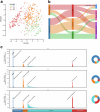
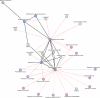
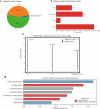
The composition of the infant gut microbiome is critical to immune development and noncommunicable disease (NCD) trajectory. However, a comprehensive evaluation of the infant gut microbiome in the United States is lacking. The My Baby Biome study, designed to address this knowledge gap, evaluated the gut microbiomes of 412 infants (representative of U.S. demographic diversity) using metagenomics and metabolomics. Regardless of birth mode and/or feeding method, widespread Bifidobacterium deficit was observed, with approximately 25% of U.S. infants lacking detectable Bifidobacterium. Bifidobacterium-dominant microbiomes exhibit distinct features when compared to microbiomes with other dominant microbial compositions including reduced antimicrobial resistance and virulence factor genes, altered carbohydrate utilization pathways, and altered metabolic signatures. In C-section birth infants, Bifidobacterium tended to be replaced in the human milk oligosaccharide utilization niche with potentially pathogenic species. Longitudinal health outcomes from these infants suggest that the disappearance of key Bifidobacterium may contribute to the development of atopy.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: Research was funded by Persephone Biosciences. All authors were employed by and/or hold stock in Persephone Biosciences. RI also holds stock in Kenvue.
Figures


Similar articles
-
The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review.BMC Gastroenterol. 2016 Jul 30;16(1):86. doi: 10.1186/s12876-016-0498-0. BMC Gastroenterol. 2016. PMID: 27475754 Free PMC article.
-
Cohort profile: Mother and Infant Metabolome and Microbiome (MIMM) study, a prospective cohort study of mothers and infants in Boston, Massachusetts.BMJ Open. 2025 Jun 18;15(6):e096957. doi: 10.1136/bmjopen-2024-096957. BMJ Open. 2025. PMID: 40533202 Free PMC article.
-
Multiple perinatal characteristics affect the association between maternal diabetes status and early neonatal gut microbiota.mSphere. 2025 Jun 25;10(6):e0091424. doi: 10.1128/msphere.00914-24. Epub 2025 May 16. mSphere. 2025. PMID: 40377305 Free PMC article.
-
Effects of maternal probiotic supplementation on breast milk microbiome and infant gut microbiome and health: a systematic review and meta-analysis of randomized controlled trials.Am J Obstet Gynecol MFM. 2023 Nov;5(11):101148. doi: 10.1016/j.ajogmf.2023.101148. Epub 2023 Sep 1. Am J Obstet Gynecol MFM. 2023. PMID: 37660760
-
Effect of Fructooligosaccharides Supplementation on the Gut Microbiota in Human: A Systematic Review and Meta-Analysis.Nutrients. 2022 Aug 12;14(16):3298. doi: 10.3390/nu14163298. Nutrients. 2022. PMID: 36014803 Free PMC article.
References
-
- Sonnenburg, E. D. & Sonnenburg, J. L. The ancestral and industrialized gut microbiota and implications for human health. Nat. Rev. Microbiol.17, 383–390 (2019). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

