Transcriptomic Profiling Reveals Potential Genes Involved in the Immune Landscape of Polycystic Ovary Syndrome: An Exploratory Study
- PMID: 40555906
- PMCID: PMC12271263
- DOI: 10.1007/s43032-025-01917-4
Transcriptomic Profiling Reveals Potential Genes Involved in the Immune Landscape of Polycystic Ovary Syndrome: An Exploratory Study
Abstract
Background: Polycystic ovary syndrome (PCOS) is a complex endocrine disorder associated with chronic inflammation, insulin resistance, and ovarian dysfunction. Emerging evidence implicates granulosa cell anoikis, a specialized form of apoptosis induced by extracellular matrix detachment, in PCOS pathogenesis. However, the involvement of anoikis-related genes (ARGs) remains poorly understood.
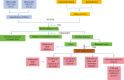
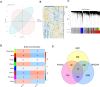
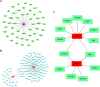
Methods: Transcriptomic data from two GEO datasets (GSE43264 and GSE98421) were integrated, followed by differential gene expression analysis and weighted gene co-expression network analysis (WGCNA). Thirteen differentially expressed ARGs (DEARGs) were identified. Functional enrichment (GO/KEGG), immune infiltration profiling (CIBERSORT), and machine learning models were used to screen for hub biomarkers. Diagnostic potential was validated using ROC and nomogram analyses. ceRNA, drug-gene, and transcription factor networks were constructed. Pancancer expression and immune relevance were assessed using TCGA, and gene function was validated through MGI.
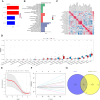
Results: Two ARGs, GSTP1 and LPCAT1, were identified as robust diagnostic markers (AUC > 0.80). These genes were significantly associated with immunosuppressive cell infiltration (e.g., elevated M2 macrophages, reduced CD8⁺ T cells). GSEA linked both genes to apoptosis, PI3K signaling, and immune pathways. MGI validation revealed that GSTP1 and LPCAT1 are involved in reproductive and metabolic regulation in murine models, supporting their functional relevance to PCOS. Drug prediction analyses identified resveratrol, curcumin, and vitamin E as potential therapeutics.
Conclusion: This is the first exploratory study to identify GSTP1 and LPCAT1 as potential diagnostic biomarkers for PCOS, validated across multi-omics platforms and functional databases. These findings highlight a novel anoikis-immunity axis in PCOS and suggest new directions for biomarker-guided therapy.
Supplementary Information: The online version contains supplementary material available at 10.1007/s43032-025-01917-4.
Keywords: Anoikis; Diagnostic biomarkers; Drug prediction; Immune landscape; Machine learning; Pancancer; Polycystic ovary syndrome.
Conflict of interest statement
Declarations. Ethics Approval and Consent to Participate: Not applicable. Consent for Pubilication: Not applicable. Conflict of Interest: The authors declare that they have no competing interests.
Figures


References
-
- Stener-Victorin E, Teede H, Norman RJ, Legro R, Goodarzi MO, Dokras A, Laven J, Hoeger K, Piltonen TT. Polycystic ovary syndrome. Nat Rev Dis Primers. 2024;10(1):27. 10.1038/s41572-024-00511-3. PMID: 38637590. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

