The genome sequence of Atlantic Bluefin Tuna, Thunnus thynnus (Linnaeus, 1758)
- PMID: 40556665
- PMCID: PMC12186022
- DOI: 10.12688/wellcomeopenres.23971.1
The genome sequence of Atlantic Bluefin Tuna, Thunnus thynnus (Linnaeus, 1758)
Abstract
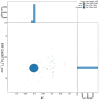
We present a genome assembly from a specimen of Thunnus thynnus (Atlantic Bluefin Tuna; Chordata; Actinopteri; Scombriformes; Scombridae). The genome sequence has a total length of 799.05 megabases. Most of the assembly (99.17%) is scaffolded into 24 chromosomal pseudomolecules. The mitochondrial genome has also been assembled, with a length of 16.53 kilobases. Gene annotation of this assembly on Ensembl identified 23,266 protein-coding genes.
Keywords: Atlantic Bluefin Tuna; Scombriformes; Thunnus thynnus; chromosomal; genome sequence.
Copyright: © 2025 Oomen RA et al.
Conflict of interest statement
No competing interests were disclosed.
Figures




References
-
- Aalto EA, Dedman S, Stokesbury MJW, et al. : Evidence of bluefin tuna ( Thunnus thynnus) spawning in the Slope Sea region of the Northwest Atlantic from electronic tags. ICES J Mar Sci. 2023;80(4):861–877. 10.1093/icesjms/fsad015 - DOI
-
- Aalto EA, Ferretti F, Lauretta MV, et al. : Stock-of-origin catch estimation of Atlantic bluefin tuna ( Thunnus thynnus) based on observed spatial distributions. Can J Fish Aquat Sci. 2021;78(8):1193–1204. 10.1139/cjfas-2019-0445 - DOI
LinkOut - more resources
Full Text Sources

